Table 1. Comparison of disorder prediction.
| Tool | TP | TN | FP | FN | accuracy | recall |
| VSL2 fast |
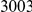
|
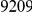
|
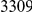
|
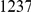
|
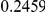
|
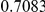
|
| VSL2 |
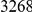
|
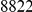
|
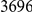
|
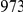
|
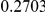
|
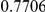
|
| iupred long |
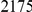
|
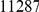
|
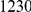
|
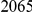
|
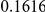
|
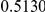
|
| iupred short |
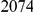
|
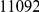
|
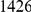
|
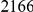
|
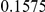
|
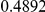
|
| phyHMM SwissProt |
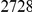
|
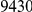
|
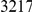
|
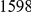
|
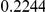
|
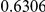
|
| phyHMM PANDIT |
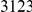
|
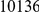
|
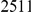
|
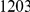
|
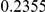
|
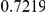
|
| HMM SwissProt |
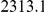
|
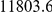
|
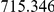
|
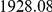
|
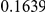
|
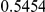
|
| HMM PANDIT |
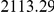
|
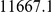
|
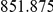
|
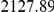
|
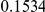
|
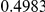
|
Comparison of phylo-HMM based disorder prediction using the models estimated from the PANDIT or the SwissProt data set with other sequence based predictors. Shown are true positives (TP), true negatives (TN), false positives (FP), false negatives (FN) for different parameter configurations of each method.
