Abstract
Little is known about the roles of aldehyde dehydrogenases in non-vertebrate animals. We recently showed that in Drosophila melanogaster, an enzyme with ~70% amino acid identity to mammalian ALDH2 is necessary for detoxification of dietary ethanol. To investigate other functions of this enzyme, DmALDH, encoded by the gene Aldh, we compared two strains homozygous for Aldh-null mutations to two closely related wild type strains in measures of fitness and stress resistance in the absence of ethanol. Aldh-null strains have lower total reproductive rate, pre-adult viability, resistance to starvation, and possibly longevity than wild-type strains. When maintained under hyperoxia, Aldh nulls die more quickly and accumulate higher levels of protein carbonyls than wild-types, thereby providing evidence that DmALDH is important for detoxifying reactive aldehydes generated by lipid peroxidation. However no effect of Aldh was seen on protein carbonyl levels in flies maintained under normoxia. It is possible that Aldh nulls experience elevated rates of protein carbonylation under normoxia, but this is compensated (at a fitness cost) by increased rates of degradation of the defective proteins. Alternatively, the fitness defects of Aldh nulls under normoxia may result from the absence of one or more other functions of DmALDH, unrelated to protection against protein carbonylation.
Keywords: protein carbonylation, Aldehyde Dehydrogenase, Drosophila, balancer equilibrium method, stress resistance
1. Introduction
Mammalian mitochondrial Aldehyde Dehydrogenase (ALDH2, EC 1.2.1.3) is well known for its role in detoxifying acetaldehyde derived from dietary ethanol [1–3]. Recently a homolog of mammalian ALDH2 was described from Drosophila melanogaster that is similarly essential for metabolizing ethanol, which can occur in significant quantities in the decaying fruit on which this species feeds. Flies lacking the enzyme, DmALDH, are killed when fed ethanol concentrations that are easily tolerated by wild-types [4]. DmALDH has a predicted mitochondrial leader sequence, and the gene encoding it, Aldh, is highly expressed in every adult and larval tissue examined [5]. The predicted protein shares a high sequence identity (60%–70%) with mammalian members of the ALDH1/ALDH2 family.
The high degree of conservation between Drosophila and mammalian ALDHs suggests that these enzymes have fundamental roles in detoxification or metabolism unrelated to catabolism of ethanol, significant quantities of which are probably not encountered by most members of the animal kingdom. Drosophila provides an excellent system for investigating these roles, not only because of its experimental tractability, but because ALDHs have undergone less extensive gene duplication in Drosophila than in mammals [6]. Notably, DmALDH has a higher amino acid identity to each of five human ALDHs (ALDH2, 1B1, 1A1, 1A2, and 1A3) than to any other ALDH in the Drosophila genome. Thus the effects of knocking out Aldh are less likely to be compensated by genes with redundant function than is the case with mammals. An additional advantage of studying Drosophila is that there is currently little information on the in vivo functions of ALDHs in any invertebrate animal.
One of the major functions of ALDH2 and some other ALDHs in mammals is believed to be detoxification of aldehydes formed by the peroxidation of polyunsaturated fatty acyl groups of phospholipids present in the mitochondrial membrane [7, 8]. These self-propagating peroxidation reactions are initiated by the hydroxyl radicals produced within the mitochondria which ultimately produce various reactive aldehydes, alkenals and hydroxyalkenals. Many of these aldehydes are cytotoxic in nature due to their reactivity towards proteins. The proteins are rendered inactive or damaged by covalent modification (carbonylation) of amino acid side chains -- for example, the sulphydryl group of cysteine, the imidazole ring of histidine and the ε-amino group of lysine -- by the aldehydes [9]. The biological effects of such damage include but may not be limited to inhibition of DNA and protein synthesis, as well as inactivation of mitochondrial enzymes such as the pyruvate dehydrogenase complex and cytochrome C oxidase, and alterations in gene expression [10–12]. Absence of ALDH2 activity leads to a higher accumulation of carbonylated proteins due to a rise in the levels of reactive aldehydes, which may lead to the development of pathological conditions [7,13]. It is possible that DmALDH, being a homolog of ALDH2, is carrying out a similar function in Drosophila.
Prompted by the above considerations, we examined whether Drosophila lacking DmALDH show defects consistent with an important role of this enzyme in the detoxification of lipid peroxidation products. First, we examined the effects of DmALDH on the overall “health” of flies by comparing reproductive rate, longevity, and starvation resistance of flies lacking DmALDH to those of closely related wild-type flies. Second, we tested whether Aldh-null flies show increased sensitivity to hyperoxia, as predicted if DmALDH is involved in detoxifying lipid peroxidation products. Finally, we compared levels of protein carbonylation in mitochondrial protein extracts of Aldh-null and wild-type flies, when reared under both normoxia and hyperoxia.
2. Materials and methods
2.1 Stocks and rearing conditions
For all experiments, we used two Aldh null mutants produced by imprecise excision of a P transposable element (lines 17 and 24 of [4]), and two control lines with precise excisions of the P-element (lines 14 and 26). The deletions in the mutant lines remove the first 41 codons of the gene, and reduce ALDH activity of whole fly homogenates with acetaldehyde as a substrate to about 10% of that of the controls [4]. The level of residual ALDH activity is similar between line 17, which produces an aberrant transcript, and line 24 [4]; this suggests that both mutants are true nulls, and that the residual activity comes from one or more other ALDHs in the genome.
We found the original lines to be difficult to rear in large numbers, in part because they had three visible markers on their second chromosomes [4]. Prior to the experiments reported here, therefore, we crossed off two of the markers, leaving only the black body color marker. This was done by crossing each line to a strain carrying a deletion encompassing Aldh and nearby genes, but lacking the markers. Chromosomes bearing only the black marker, but lacking the deletion (as assessed by homozygous viability) were recovered and used to establish three sublines from each line, each derived from a different recombination event. These sublines were used shortly thereafter for the starvation resistance experiment. Because the genetic backgrounds (first, third and fourth chromosomes) of these original sublines contained a mix of material from the stocks used to construct them, we subsequently replaced their first and third chromosomes with those from a balancer stock bearing the visible marker veinlet on the third chromosome [14] (the small fourth chromosome was not controlled in these crosses, but comprises <1% of the D. melanogaster genome). These more genetically homogeneous sublines were used for all the other experiments described here.
Flies were reared in shell vials containing approximately 10 ml of cornmeal-molasses-dead yeast medium, and handled under light CO2 anesthesia. Except for the hyperoxia experiments, flies were maintained in a walk-in environmental chamber at 25°C and 60–70% relative humidity, under continuous lighting. The hyperoxia experiments were conducted on a laboratory bench at 21–23°C.
2.2 Total fitness and pre-adult viability
We used the balancer-equilibrium method of Sved and Ayala [15] to compare total fitness (reproductive success, averaged over sexes, including the effect of pre-reproduction mortality) of Aldh-null and control genotypes when reared under crowded conditions. This method takes advantage of balancers, chromosomes with multiple inversions that effectively suppress recombination with non-inverted homologues, one or more dominant visible markers, and one or more recessive lethals. If “Bal” denotes a balancer chromosome and “+i” a single non-inverted homologue, populations started from Bal/+i flies will usually come to an equilibrium in which Bal/+i and +i/+i flies are present in a characteristic ratio. This occurs because the heterozygous (Bal/+i) genotype has higher fitness than either homozygous genotype: the Bal/Bal combination is lethal, while the +i/+i combination creates homozygosity for deleterious recessives that are present on all wild-type chromosomes. The relative proportions of Bal/+i and +i/+i in zygotes at equilibrium can be used to calculate the fitness of the +i/+i genotype as a proportion of that of the Bal/+i genotype. Furthermore, because most mutations have much larger fitness effects as homozygotes than as heterozygotes, the fitness of balancer heterozygotes with different + chromosomes (e.g., Bal/+i and Bal/+j) can be safely regarded as roughly equivalent. Thus, a difference in the relative fitness of two +/+ genotypes is likely to stem largely from a difference in their absolute fitness.
We used the CyO balancer, containing the dominant markers Curly (wings) and Rough eye. To establish each population, CyO/+ijk females were crossed to CyO/+ijk′ males; here, i represents Aldh genotype (mutant or wild-type), j represents line (17 or 24 if mutant, 14 or 26 if wild-type), and k represents subline within line (1, 2 or 3), with k ≠ k′. This was done for each of the six possible pairs of sublines within lines, creating a total of 24 populations, divided equally into two blocks initiated one week apart (crosses used to establish the second block were the reciprocals of those used to establish the first block; one population was later discarded due to contamination). We chose to establish populations by hybridizing sublines in order to minimize the effects of random deleterious mutations unique to each subline. This choice has the disadvantage, though, of creating non-independence between the populations derived from the same line. As a result, statistical analyses are based on means of the populations for a given line.
Each initial cross consisted of six vials with six females and 4–5 males per vial. Flies were allowed to lay eggs for three days and were then removed. Thirteen days after the vials were set up, emerged Curly and non-Curly flies were counted, and flies from all six vials were mixed in an empty bottle. Approximately 40 flies from the bottle were then distributed, irrespective of genotype, into each of six holding vials, with leftover flies discarded. After one day of feeding and mating, flies in the holding vials were transferred to new vials to lay eggs for the next generation. This cycle was repeated for nine generations, by which time the frequencies of Curly and non-Curly flies had roughly stabilized.
To estimate relative fitness of the +/+ flies (note that “+” in this context refers to the absence of the balancer, not Aldh genotype), it is necessary to estimate the equilibrium zygotic frequencies of the Cy/+ and +/+ genotypes, which are likely to differ from the frequencies among adults [16]. This was done by allowing flies from the final generation of each population to lay eggs at a low density (10 pairs of flies in each of three half-pint bottles containing 30 ml of food, with +/+ and Cy/+ flies chosen in proportion to their abundance in the population), with the goal of reducing differential mortality before progeny were scored. Because some differential mortality could still have occurred under these conditions, we separately estimated the relative survival of +/+ to Cy/+ genotypes for each population by setting up similar bottles with only Cy/+ males and virgin females. Emergees from both sets of bottles were counted up until day 17, by which time almost all flies had emerged. As shown by Sved [16], if the proportions of Curly flies among the emergees from the first and second type of bottles are denoted by h and r, respectively, the relative fitness of +/+ to Cy/+ flies can be estimated as (r – h)/[r (1 – h)].
Although the main goal of this experiment was to compare the total fitness of Aldh-null and wild-type flies, the proportions of Curly flies among the emergees from the founding crosses and from the Cy/+ × Cy/+ crosses at the final generation can be used to compare pre-adult viability of Aldh-nulls and wild-types at medium and low larval densities, respectively. In the absence of differential mortality, both crosses would be expected to produce a ratio of one +/+: two Cy/+ among the adult progeny; thus the survival of +/+ relative to Cy/+ larvae can be estimated as twice the ratio of non-Curly to Curly emergees. In calculating the final generation viability estimates, we ignored flies which emerged after day 13, in order to make these estimates more directly comparable to those from the first generation. Because not all flies had emerged by day 13 in either case (although most had), these viability estimates may partly reflect differences in development time, with slower development appearing as lower viability.
2.3 Starvation resistance, longevity, and hyperoxia resistance
Starvation resistance, longevity, and hyperoxia resistance were measured on male flies held in groups of ten per vial. To measure starvation resistance, 3–8 day old flies were placed in vials containing a small cotton ball moistened with two ml of distilled water. Six such vials were set up per subline, and mortality was scored every 12 hours until all flies had died. Longevity was measured by placing newly-emerged males in vials containing normal medium, with eight vials per subline; flies were transferred without anesthesia to new vials every three days and mortality scored daily until all flies had died.
Hyperoxia resistance was measured by placing 0–2 day old flies in vials with medium, with 2–4 vials per subline. One day later, the vials were placed inside a glove bag (Glas-Col, Terre Haute, IN) with an inlet tube through which 100% oxygen entered from a tank. After the bag inflated, oxygen concentration, as determined by an oxygen meter (Extech Instruments, Waltham, MA), was maintained in the range 90–100%. Flies were transferred to vials with fresh medium after the second day and survival recorded after the third day.
2.4 Protein carbonylation assay
Total carbonylated protein content was measured on protein extracts from male flies using 2, 4-dinitrophenylhydrazine [17]. The extraction procedure used was designed to enrich the mitochondrial protein fraction, although some proteins from other sources may have been present. Flies (100/sample) were homogenized in 1 ml isolation buffer (.25M sucrose, 10mM Tris, pH 7.4, 1 mM EDTA, 1% BSA) with protease inhibitor cocktail (Roche Complete Mini Protease Inhibitor, Roche Applied Science). The homogenized fly extract was filtered through an 11μm pore size nylon membrane (Millipore, cat. no. NY1104700) and the filtrate was centrifuged at 150g for five minutes. The supernatant was saved and streptomycin sulfate solution was added to a final concentration of 1%. After incubating for 15 minutes the extract was centrifuged at 11,000g for ten minutes. A 200 ul aliquot of supernatant was placed in each of two 2 ml microcentrifuge tubes; 800ul of 10mM DNPH (Sigma-Aldrich) solution in 2M HCl was added to one tube (sample) and 800ul 2M HCl was added to the other (standard). Both standard and sample tubes were incubated for one hour at room temperature in the dark with occasional mixing, after which 1 ml 20% trichloroacetic acid was added to precipitate proteins. Sample and standard tubes were centrifuged at 11,000g for 3 minutes at 4°C. The supernatant was discarded and the pellets were washed 5 times with 1 ml of ethanol: ethyl acetate (1:1, v/v). After the final wash, the pellet was dissolved in 600ul of guanidinium hydrochloride by vortexing thoroughly and then incubating at 37°C for 15 minutes. Any insoluble particle was precipitated by centrifuging the solution at 11,000g for three minutes. The absorbance of the supernatant was read at 375 nm. The difference in absorbance values between the sample and the standard were converted to carbonyl concentration using the molar absorbing coefficient (ε = 22000 M−1 cm−1) of the hydrazones and a cuvette pathlength of 1 cm.
In experiment one, flies were exposed to hyperoxia for two days before extraction. In experiment two, performed at a different time, flies were kept under normoxic conditions. In each case, a single extraction was performed on each subline. Experiment 3 was designed to remedy two shortcomings of the first two experiments. First, to allow a direct comparison of carbonylation levels between oxygen treatments, the two treatments were set up simultaneously, and flies from each treatment processed at the same time. Second, protein concentration of the final supernatant was estimated with a Qubit fluorometer (Invitrogen) and was used to normalize carbonyl concentration of each sample. Flies for this experiment were F1 hybrids from crosses of a pair of sublines from each line; thus there were two mutant and two control samples per oxygen treatment.
2.5 Statistical analysis
Statistical analysis was carried out using SAS version 9.2. Line means for fitness and viability were compared between line types (Aldh-null or control) by two-sample t-tests. For starvation resistance and longevity, mean and median survival times per vial, respectively, were analyzed using the MIXED procedure (see [18] for details), with sublines nested within lines (both random effects), and lines nested within line type (a fixed effect). For hyperoxia resistance, a similar analysis was performed on the arcsin square-root transformed proportions of flies surviving per vial. Carbonylation levels from experiments 1 and 2 were analyzed in similar fashion, except no replication was present at the level of sublines. For experiment 3, a two-way analysis was performed, with treatment (hyperoxia or normoxia) crossed with line type, and line nested within line type. Because a specific direction of difference was expected in all comparisons of mutants and controls (e.g., lower viability and fitness of the former), one-tailed P values are reported.
3. Results
3.1 Aldh-null flies are impaired in total fitness, preadult viability, and starvation resistance
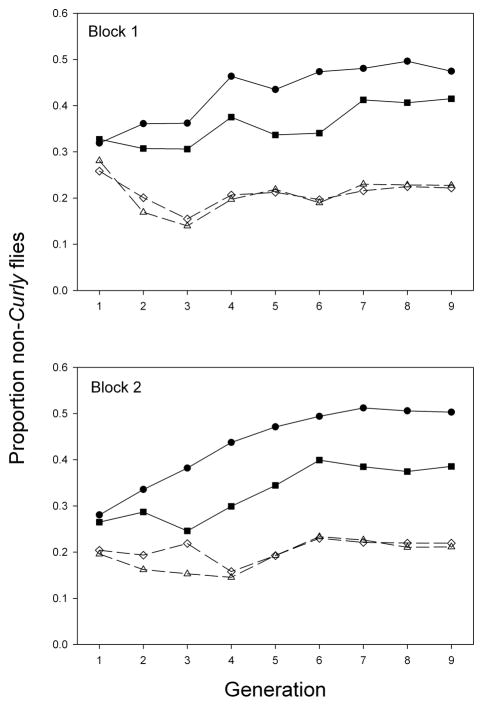
To determine the effects of Aldh loss of function mutations on total fitness, populations were established in which Aldh+ and Aldh− chromosomal homozygotes competed against Curly balancer heterozygotes, which always have a functional Aldh copy provided by the balancer chromosome. Non-Curly flies rose to higher frequencies in populations in which they were homozygous for a functional Aldh copy than in populations in which they were homozygous for an Aldh-null mutation (Fig. 1, closed and open symbols, respectively). In both blocks, frequencies of non-Curly flies roughly stabilized by generation 7 (Fig. 1). The total fitness of non-Curly homozygotes relative to balancer heterozygotes, estimated at the last generation (see Materials and Methods), was on average 2.75 times higher in the Aldh+ lines than in the Aldh− lines (Table 1, last column). Thus, although Aldh− homozygotes are qualitatively viable and fertile in both sexes, they had strongly reduced reproductive rates compared to Aldh+ homozygotes under the crowded conditions of the experimental populations. Aldh+ homozygotes also had significantly higher pre-adult viability than Aldh− homozgyotes at both low and medium larval densities, although the differences were not nearly as pronounced as for total fitness (Table 1).
Fig. 1.
Results of balancer-equilibrium experiment, showing mean proportions of Aldh+ (solid lines and closed symbols) or Aldh– (dashed lines and open symbols) chromosomal homozygotes among adults. Both genotypes competed with Curly balancer heterozygotes, which have at least one functional Aldh copy. Circles, line 14; squares, line 26; triangles, line 17; diamonds, line 24. Each point is the mean of three populations, with >500 flies counted per population most generations.
Table 1.
Egg-to-adult viability and total fitness of Aldh null mutant and control lines from the balancer equilibrium experiment.
| Egg-to-adult viability | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type | Line | Low larval density | Medium larval density | Total fitness | ||||||
| Block 1 | Block 2 | Mean | Block 1 | Block 2 | Mean | Block 1 | Block 2 | Mean | ||
| Control | 14 | 0.94 | 1.04 | 0.99 | 0.94 | 0.78 | 0.86 | 0.61 | 0.62 | 0.62 |
| 26 | 0.93 | 1.13 | 1.03 | 0.97 | 0.73 | 0.85 | 0.53 | 0.43 | 0.48 | |
| Mutant | 17 | 0.83 | 0.92 | 0.87 | 0.78 | 0.49 | 0.64 | 0.31 | 0.20 | 0.26 |
| 24 | 1.02 | 0.80 | 0.91 | 0.70 | 0.51 | 0.60 | 0.04 | 0.24 | 0.14 | |
| Prob. | 0.023 | 0.002 | 0.028 | |||||||
Values are relative to the Curly balancer genotype in each population; this genotype has a wild-type Aldh. Probabilities are from one-tailed t-tests.
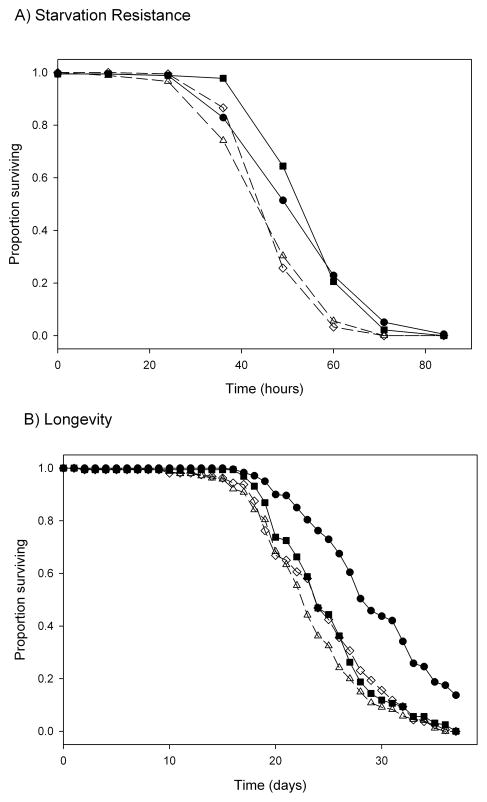
Aldh-null homozgyotes died more quickly in the absence of food than Aldh+ homozygotes (Fig. 2A); mean survival time was 49.5 hours in the mutant lines and 57.0 hours in the controls (P < 0.03). There was no significant effect of line within line type (mutant or control) on starvation resistance (P > 0.5). The results for longevity were more ambiguous: one wild-type line survived longer than both the two mutant lines and the other wild-type line, which differed little from each other in longevity (Fig. 2B). In this experiment, there was no significant effect of line type on longevity (P = 0.095), but there was a significant effect of line within line type (P = 0.03). The wild-type line with low longevity also had lower fitness than the other wild-type line in the balancer equilibrium experiment (Fig. 1, Table 1). This suggests that this line carries a fitness-reducing mutation at another gene; such cryptic mutations sometimes occur when P elements are mobilized [19].
Fig. 2.
Starvation resistance (A) and longevity (B) of Aldh+ and Aldh– homozygous flies. Line symbols are as in Fig. 1.
3.2 Aldh-null flies are sensitive to hyperoxia, and accumulate elevated levels of protein carbonyls under hyperoxia, but not normoxia
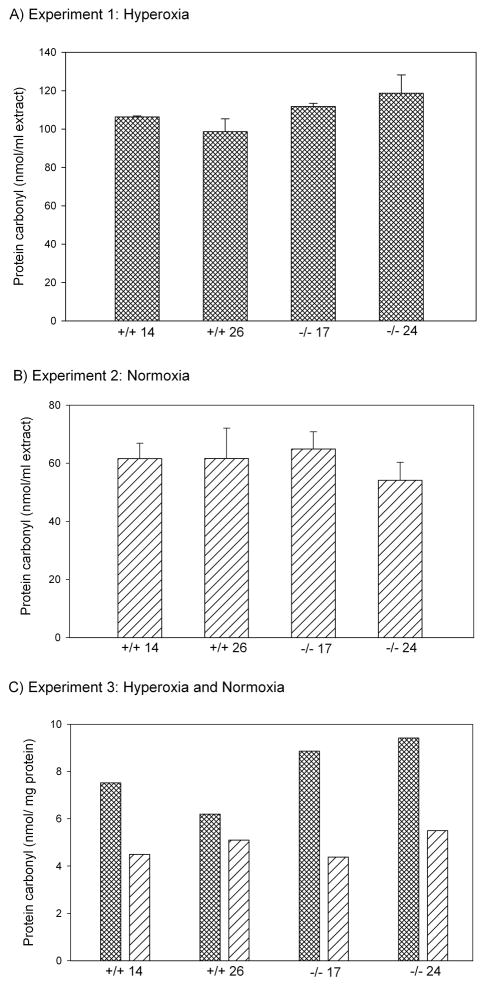
When maintained in an atmosphere containing >90% oxygen, the Aldh-null lines experienced markedly higher mortality after three days than the control lines (Fig. 3; line type, P < 0.025; line within type, P > 0.1). Replicate hyperoxia resistance experiments gave similar results (data not shown). To determine whether the hyperoxia sensitivity of mutants might be caused by higher levels of protein carbonylation than controls, carbonylation was measured in protein extracts of flies exposed to hyperoxia for two days, before mortality had occurred. Extracts from mutants had significantly higher levels of protein carbonyls than extracts from controls (Fig. 4A; line type, P < 0.03; line within type, P > 0.5).
Fig. 3.
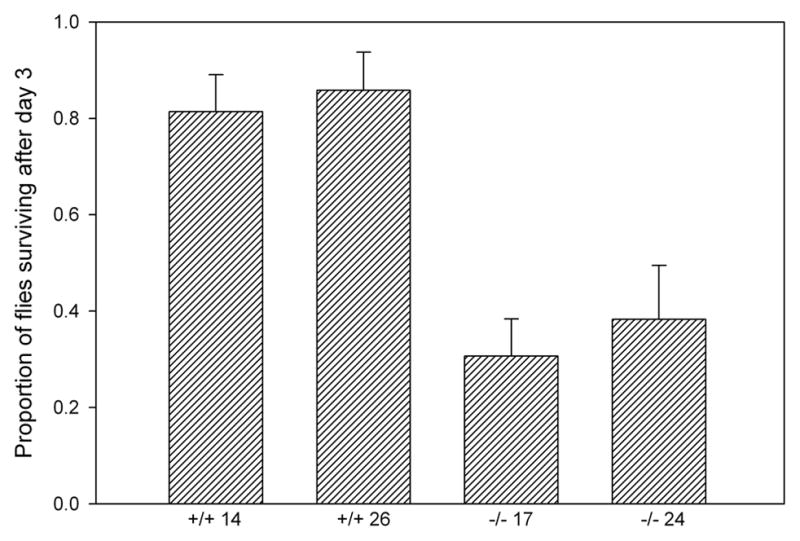
Survival under hyperoxia of Aldh+ and Aldh– homozygous flies. Bars are standard errors among sublines.
Fig. 4.
Protein carbonylation levels of of Aldh+ and Aldh– homozygous flies under (A) hyperoxia, (B) normoxia, and (C) in a replicate experiment comparing the two treatments simultaneously. Bars in (A) and (B) are standard errors among sublines; (C) shows results of single samples of hybrids between two sublines per line.
Under normoxia, in contrast, there was no difference between Aldh-null mutants and controls in protein carbonylation levels (Fig. 4B; line type, P > 0.5; line within type, P > 0.5). Mean carbonylation levels were lower than in the experiment with hyperoxia, consistent with the expectation that hyperoxia increases lipid peroxidation. To check these results, a third experiment was carried out in which oxygen treatments, protein extraction, and carbonylation measurement were carried out simultaneously, and carbonyl concentrations normalized to the protein concentration of the extracts (Fig. 4C). Confirming the results of the first two experiments, hyperoxia exposure significantly increased levels of protein carbonyls (P < 0.003), but did so more in Aldh-null flies than in controls (interaction between line type and oxygen treatment, one-tailed P < 0.05; a one-tailed test is justified because the observed direction of interaction is predicted by the results of the first two experiments). Under normoxic conditions, there was again no difference in protein carbonyl concentration between Aldh-null and control flies (Fig. 4C).
4. Discussion
In the absence of ethanol, flies lacking DmALDH, a member of the ALDH1/2 family, have strongly reduced reproductive rate and moderately reduced preadult viability compared to genetically-matched control flies. Lack of DmALDH also impairs starvation resistance, and may reduce longevity, although the difference between the two control lines rendered the longevity experiment ambiguous. These defects support an important role of DmALDH in functions unrelated to ethanol metabolism, consistent with the high degree of conservation of the protein between flies and mammals.
The null mutants also experience higher mortality under hyperoxia compared to controls, as would be expected if DmALDH detoxifies reactive aldehydes resulting from lipid peroxidation. Consistent with this hypothesis, under hyperoxia, Aldh-null mutants accumulate higher levels of protein carbonyls than controls. Surprisingly, no difference in levels of protein carbonyls was observed between mutants and controls under normoxic conditions. This negative result raises the question of whether the fitness defects of Aldh-null mutants under normoxia have anything to do with protection against protein carbonylation by reactive aldehydes, or whether there is an additional important function of DmALDH. One possibility is that under normoxic conditions, Aldh-null mutants experience higher rates of protein carbonylation, but this is compensated by higher rates of proteosomal degradation of the carbonylated proteins [20–22], resulting in similar steady-state levels of protein carbonyls in mutants and controls. Higher protein turnover would nonetheless be energetically costly, thus accounting for the fitness defects (e.g., lower starvation resistance) of the null mutants. Under hyperoxia, in contrast, the protein degradation pathway could become overburdened in the mutants and proteins involved in degradation may be inhibited [22], resulting in higher levels of protein carbonyls than in controls. This hypothesis could be tested by inhibiting protein degradation and/or measuring protein turnover in Aldh-null mutants and controls under normoxia [23].
A carbonyl reductase [24] and glutathione S transferase [25] in D. melanogaster have also been shown to protect cells from reactive aldehydes. It is possible that in Aldh-null mutants, these enzymes functionally compensate for the absence of DmALDH but probably at a cost of impairing their other functions. Such functional compensation might maintain effective protein carbonylation in the mutants at a level similar to that of the controls with a reduction in overall fitness of the mutants due to the cost of the compensation. Under hyperoxia, in contrast, such compensatory contributions from the other enzymes might not be sufficient to regulate the excessive amount of reactive aldehydes produced leading to higher accumulation of protein carbonyls in mutants compared to the control flies. This hypothesis could be investigated by testing the activities of the abovementioned enzymes in Aldh-null mutants.
A third possibility is that DmALDH has a role in metabolism in addition to detoxification of the reactive aldehydes responsible for most protein carbonylation. For example, the retinoic acid (RA) signaling pathway appears to be important in some aspects of Drosophila development [26] and RA is known to provide protection from oxidative stress in mammals [27]. Therefore DmALDH could potentially serve as a retinaldehyde dehydrogenase, like some members of the ALDH1/2 family in mammals [28]. It is not clear, however, whether impaired RA signaling would be sufficient to account for the fitness defects of Aldh-null flies. We are also skeptical of the results of Halme et al. [26] who suggested that DmALDH plays a role in RA signaling, because the putative Aldh mutant that they used had ALDH activity in the wild-type range when we tested it [4]. This line was the precursor of our mutant and control lines.
Rothacker and Ilg [29] examined the activity of DmALDH with a variety of aldehydes, showing that it is an effective catalyst with acetaldehyde and short chain aliphatic aldehydes through hexanal. However the reaction rate dropped off markedly when octanal was used as a substrate. They used an uncommon variant of the enzyme, however, with Phe substituted for the ancestral Leu at position 479. The Phe479 variant is in low frequency in natural populations [30], and shifts substrate specificity towards shorter chain aldehydes ([30]; Chakraborty and Fry, unpublished data). We are currently characterizing substrate specificity of the Leu479 and Phe479 variants in detail, and carrying out additional experiments to investigate the in vivo function of DmALDH.
Acknowledgments
This work was supported by NSF (DEB-0623268) and NIH (R01AA016178) grants to J.D.F. Travel of M.C. to present this work at the 15th IMEMBCM in Lexington, KY USA was supported by USPHS NIH grant R13-AA019612. We thank L. Harshman and E. Burr for doing preliminary hyperoxia experiments, J. Budnik for carrying out the balancer equilibrium experiment, D. Loehlin for collecting the starvation resistance data, and K. Donlon for general laboratory assistance. J.D.F. is especially indebted to Dr. Henry Weiner for introducing him to the carbonyl metabolism community.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eckfeldt J, Mope L, Takio K, Yonetani T. Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes. J Biol Chem. 1976;251:236–240. [PubMed] [Google Scholar]
- 2.Greenfield NJ, Pietruszko R. Two aldehyde dehydrogenases from human liver. Isolation via affinity chromatography and characterization of the isozymes. Biochim Biophys Acta. 1977;483:35–45. doi: 10.1016/0005-2744(77)90005-5. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci U S A. 1984;81:258–261. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fry JD, Saweikis M. Aldehyde dehydrogenase is essential for both adult and larval ethanol resistance in Drosophila melanogaster. Genet Res. 2006;87:87–92. doi: 10.1017/S0016672306008032. [DOI] [PubMed] [Google Scholar]
- 5.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sophos NA, Vasiliou V. Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem Biol Interact. 2003;143–144:5–22. doi: 10.1016/s0009-2797(02)00163-1. [DOI] [PubMed] [Google Scholar]
- 7.Ohta S, Ohsawa I, Kamino K, Ando F, Shimokata H. Mitochondrial ALDH2 deficiency as an oxidative stress. Ann N Y Acad Sci. 2004;1011:36–44. doi: 10.1007/978-3-662-41088-2_4. [DOI] [PubMed] [Google Scholar]
- 8.Ohsawa I, Nishimaki K, Yasuda C, Kamino K, Ohta S. Deficiency in a mitochondrial aldehyde dehydrogenase increases vulnerability to oxidative stress in PC12 cells. J Neurochem. 2003;84:1110–1117. doi: 10.1046/j.1471-4159.2003.01619.x. [DOI] [PubMed] [Google Scholar]
- 9.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 10.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 11.Humphries KM, Yoo Y, Szweda LI. Inhibition of NADH-linked mitochondrial respiration by 4-hydroxy-2-nonenal. Biochemistry. 1998;37:552–557. doi: 10.1021/bi971958i. [DOI] [PubMed] [Google Scholar]
- 12.Musatov A, Carroll CA, Liu YC, Henderson GI, Weintraub ST, Robinson NC. Identification of bovine heart cytochrome c oxidase subunits modified by the lipid peroxidation product 4-hydroxy-2-nonenal. Biochemistry. 2002;41:8212–8220. doi: 10.1021/bi025896u. [DOI] [PubMed] [Google Scholar]
- 13.Wang B, Wang J, Zhou S, Tan S, He X, Yang Z, Xie YC, Li S, Zheng C, Ma X. The association of mitochondrial aldehyde dehydrogenase gene (ALDH2) polymorphism with susceptibility to late-onset Alzheimer’s disease in Chinese. J Neurol Sci. 2008;268:172–175. doi: 10.1016/j.jns.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Fry JD, Keightley PD, Heinsohn SL, Nuzhdin SV. New estimates of the rates and effects of mildly deleterious mutation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1999;96:574–579. doi: 10.1073/pnas.96.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sved JA, Ayala FJ. A population cage test for heterosis in Drosophila pseudoobscura. Genetics. 1970;66:97–113. doi: 10.1093/genetics/66.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sved JA. An estimate of heterosis in Drosophila melanogaster. Genet Res. 1971;18:97–105. doi: 10.1017/s0016672300012453. [DOI] [PubMed] [Google Scholar]
- 17.Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz AG, Ahn BW, Shaltiel S, Stadtman ER. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 18.Fry JD. Direct and correlated responses to selection for larval ethanol tolerance in Drosophila melanogaster. J Evol Biol. 2001;14:296–309. [Google Scholar]
- 19.Hiesinger PR, Bellen HJ. Flying in the face of total disruption. Nat Genet. 2004;36:211–212. doi: 10.1038/ng0304-211. [DOI] [PubMed] [Google Scholar]
- 20.Rivett AJ. Purification of a liver alkaline protease which degrades oxidatively modified glutamine synthetase. Characterization as a high molecular weight cysteine proteinase. J Biol Chem. 1985;260:12600–12606. [PubMed] [Google Scholar]
- 21.Marcillat O, Zhang Y, Lin SW, Davies KJ. Mitochondria contain a proteolytic system which can recognize and degrade oxidatively-denatured proteins. Biochem J. 1988;254:677–683. doi: 10.1042/bj2540677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 23.Grune T, Reinheckel T, Joshi M, Davies KJ. Proteolysis in cultured liver epithelial cells during oxidative stress. Role of the multicatalytic proteinase complex, proteasome. J Biol Chem. 1995;270:2344–2351. doi: 10.1074/jbc.270.5.2344. [DOI] [PubMed] [Google Scholar]
- 24.Botella JA, Ulschmid JK, Gruenewald C, Moehle C, Kretzschmar D, Becker K, Schneuwly S. The Drosophila Carbonyl Reductase Sniffer Prevents Oxidative Stress-Induced Neurodegeneration. Curr Biol. 2004;14:782–786. doi: 10.1016/j.cub.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 25.Singh SP, Coronella JA, Benes H, Cochrane BJ, Zimniak P. Catalytic function of Drosophila melanogaster glutathione S-transferase DmGSTS1–1 (GST-2) in conjugation of lipid peroxidation end products. Eur J Biochem. 2001;268:2912–2923. doi: 10.1046/j.1432-1327.2001.02179.x. [DOI] [PubMed] [Google Scholar]
- 26.Halme A, Cheng M, Hariharan IK. Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr Biol. 2010;20:458–463. doi: 10.1016/j.cub.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitamura M, Ishikawa Y, Moreno-Manzano V, Xu QH, Konta T, Lucio-Cazana J, Furusu A, Nakayama K. Intervention by retinoic acid in oxidative stress-induced apoptosis. Nephrol Dial Transplant. 2002;17:84–87. doi: 10.1093/ndt/17.suppl_9.84. [DOI] [PubMed] [Google Scholar]
- 28.Zhao D, McCaffery P, Ivins KJ, Neve RL, Hogan P, Chin WW, Drager UC. Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur J Biochem. 1996;240:15–22. doi: 10.1111/j.1432-1033.1996.0015h.x. [DOI] [PubMed] [Google Scholar]
- 29.Rothacker B, Ilg T. Functional characterization of a Drosophila melanogaster succinic semialdehyde dehydrogenase and a non-specific aldehyde dehydrogenase. Insect Biochem Mol Biol. 2008;38:354–366. doi: 10.1016/j.ibmb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Fry JD, Donlon K, Saweikis M. A worldwide polymorphism in aldehyde dehydrogenase in Drosophila melanogaster: evidence for selection mediated by dietary ethanol. Evolution. 2008;62:66–75. doi: 10.1111/j.1558-5646.2007.00288.x. [DOI] [PubMed] [Google Scholar]