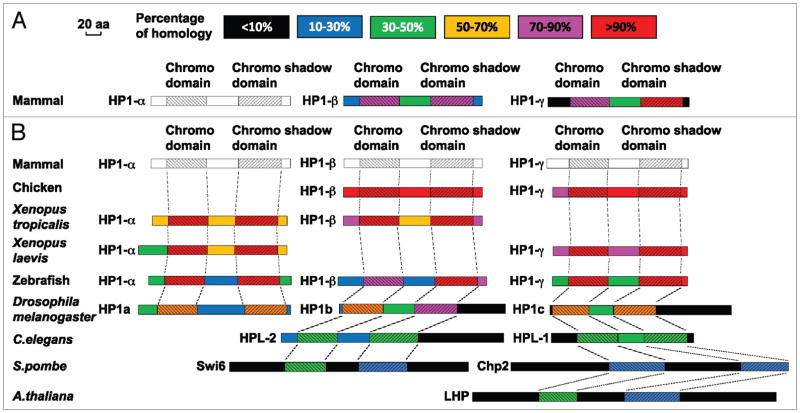
Figure 1.
Alignment of HP1 amino acid sequences. All available HP1 amino acid sequences are from the NCBI protein sequence database. In the alignment results, besides the chromo domain (CD) and the chromo shadow domain (CSD), the N-terminus adjacent to the CD, the C-terminus adjacent to the CSD, and the middle fragment between the two domains (i.e., the linker/hinge domain) are compared between the different proteins. Therefore, each protein diagram is separated into five fragments and in each fragment, the degree of homology is indicated with different colors. The length of each diagram is proportional to the protein size. (A) Alignment of mammalian HP1 amino acid sequences. The sequences of mammalian HP1β and HP1γ are compared with HP1α. The percentage of homology is based on comparison to HP1α (indicated in white/gray). (B) Alignment of HP1 amino acid sequences between different species. The HP1 amino acid sequences from several non-mammalian species are compared to the corresponding mammalian HP1 variant and their homology with mammalian HP1 is indicated in the color diagram. For C. elegans, S. pombe and A. thaliana, the homology between their HP1 proteins and each individual mammalian HP1 variant is low. Therefore, in the diagram, their sequences are compared to the mammalian HP1 variant with the highest alignment score. The alignment of the chromo and chromo shadow domains is delineated with dashed lines. Since all three mammalian HP1 sequences are used as standards for the individual alignments, they are all shown in white/gray.