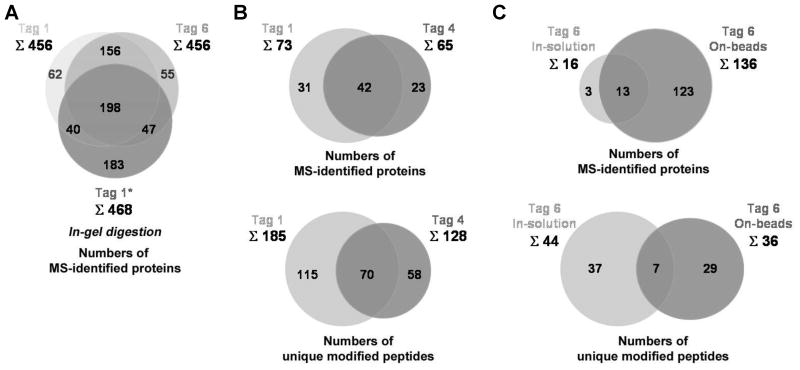
Figure 6. Overview of the proteomic analysis of AOA/ANL-labeled S. typhimurium proteins using diazobenzene-based cleavable affinity tags.
(A) Comparison of tag 1 and tag 6 in the numbers of their MS-identified AOA-labeled proteins via in-gel digestion approach and the comparison of these data to the previously published data (tag 1*) (Grammel et al., 2010). Starting with 4 mg of 1 mM Met/AOA-metabolically labeled cell lysates, the in-gel digestion approach gave total 456 protein hits for either tag 1 or tag 6. (B) Comparison of tag 1 and tag 4 in the numbers of their MS-identified AOA- and ANL-metabolically labeled proteins and unique modified peptides via in-solution digestion approach. Starting with 10 mg of 1 mM Met/AOA and Met/ANL-metabolically labeled cell lysates, the in-solution digestion approach yields total 185 unique modified peptides accounted for 73 proteins for tag 1 and total 128 unique modified peptides accounted for 65 proteins for tag 4. (C) Comparison of on-beads digestion approach and in-solution digestion approach in the numbers of their MS-identified AOA-metabolically labeled proteins and unique modified peptides using 6 as the affinity tag. Starting with 2 mg of 1 mM Met/AOA-metabolically labeled cell lysates, the in-solution digestion approach yields total 44 unique modified peptides accounted for 16 proteins whereas the on-beads digestion approach yields total 136 proteins as well as 36 unique modified peptides. See also Figure S5D and Tables S1–S5.