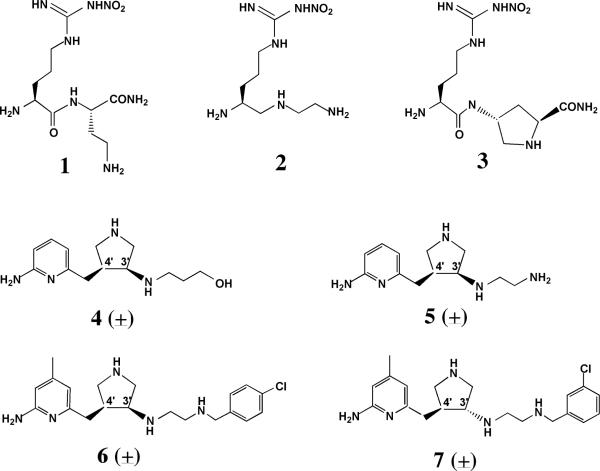
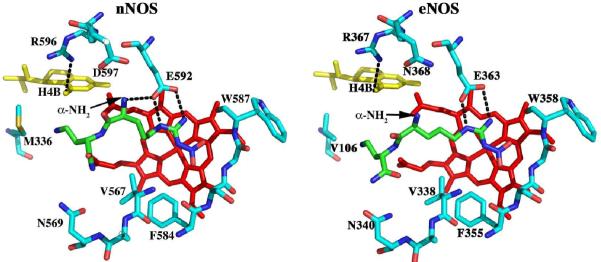
Fig. 1.
A) Chemical structures and nomenclature for the inhibitors discussed in the paper. 1. L-Nω-nitroarginine-2,4-L-diaminobutyramide; 2. (4S)-N-(4-amino-5-[aminoethyl]aminopentyl)-N'-nitroguanidine; 3. L-Nω-nitroarginine-(4R)-amino-L-proline amide; 4. (±)-3-{cis-4'-[(6“-aminopyridin-2”-yl)methyl]pyrrolidin-3'-ylamino}propan-1-ol; 5. (±)-N1-{cis-4'-[(6“-aminopyridin-2”-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine; 6. (±)-N1-{cis-4'-[(6“-amino-4”-methylpyridin-2“-yl)methyl]pyrrolidin-3'-yl}-N2-(4'-chlorobenzyl)ethane-1,2-diamine; 7. (±)-N1-{trans-4'-[(6“-amino-4”-methylpyridin-2”-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-chlorobenzyl)ethane-1,2-diamine. B) Structure of 1 complexed to nNOS and eNOS12. In nNOS the inhibitor adopts a curled conformation in order to enable the inhibitor α-amino group to optimally interact with both Glu592 and Asp597. In eNOS the residue corresponding to Asp597 is Asn368 and as a result, the inhibitor adopts an extended conformation.