Abstract
This is the first report of avian-like H6N6 swine influenza virus from swine in southern China. Phylogenetic analysis indicated that this virus might originate from domestic ducks. Serological surveillance suggested there had been sporadic H6 swine influenza infections in this area. Continuing study is required to determine if this virus could be established in the swine population and pose potential threats to public health.
Keywords: H6N6, swine influenza virus, influenza A virus, influenza pandemic
Influenza A virus can infect various hosts, including migratory waterfowl, resident birds, horses, swine, dogs, sea mammals, and humans. Among these species, swine serve as an important component of the human-animal interface and play an important role in influenza evolution and ecology. The pig is proposed as a “mixing vessel” and an intermediate host for generating novel influenza reassortants including 1968, 1957, and 2009 pandemic influenza viruses (Scholtissek, 1990). The 2009 H1N1 pandemic virus is a swine-origin reassortant: HA, NP and NS segments from classical swine (North American) lineage; PB2 and PA segments from avian (North American) lineage; PB1 segment from human seasonal H3N2; and NA and MP segments from Eurasian swine lineage (Dawood et al., 2009).
H6 subtype of influenza A virus has been identified widely in migratory waterfowl (Munster et al., 2007) as well as the resident birds such as chicken, quail, pheasant, domestic geese and ducks (Chin et al., 2002; Lee et al., 2006; Woolcock et al., 2003). Experimental infection demonstrated that H6 avian influenza virus (AIV) can replicate in the upper respiratory track and cause mild clinical symptoms in humans (Beare and Webster, 1991). Serological surveillance suggested that H6 AIVs could infect veterinarians exposed to domestic birds (Myers et al., 2007). This subtype caused also an outbreak in chicken farms in South Africa (Abolnik et al., 2007). In southern China, new lineages of H6 AIVs have been circulating in the domestic duck (Cheung et al., 2007; Chin et al., 2002; Huang et al., 2010). We here report an H6N6 swine influenza virus (SIV) from sick pigs on a small farm in southern China, and our serological surveillance suggested there were sporadic H6 swine influenza infections at four swine farms in this area. The results from this study indicate that H6 influenza A virus could use swine as an intermediate host before adapting to and causing infections in humans.
Southern China is designated as a putative influenza epicenter (Shortridge and Stuart-Harris, 1982). In order to monitor the epidemics of SIVs and to identify the potential swine influenza threats in southern China, we performed an active surveillance in the swine population in southern China. A total number of 847 samples were collected from 23 swine farms across Guangdong, Guangxi and Hainan Provinces, China. These samples include nasal swabs and lungs from sick pigs showing symptoms of coughing and/or rhinorrhea. Among these samples, thirty-five were shown to be RT-PCR positive for influenza A viruses. The RT-PCR tests were conducted based on the WHO Manual on Animal Influenza Diagnosis and Surveillance. Both nasal swabs and lung tissue homogenate were inoculated into 10-day-old specific pathogen free embryonated chicken egg, through the allantoic route, and incubated at 35 °C for 72 h. Subtype identification was conducted through standard hemagglutination inhibition and neuraminidase inhibition assays. Nine strains of SIVs were isolated: two H1N1, one H1N2, one H9N2, four H3N2, and one H6N6.
Due to the potential threats of H6 influenza A virus to public health, this study focused on the H6N6 subtype, A/swine/Guangdong/K6/2010 (H6N6). The genome of this H6N6 SIV was sequenced fully as described previously (Li et al., 2010), with the GenBank accession numbers HM800947 and HM804474 to HM804480.
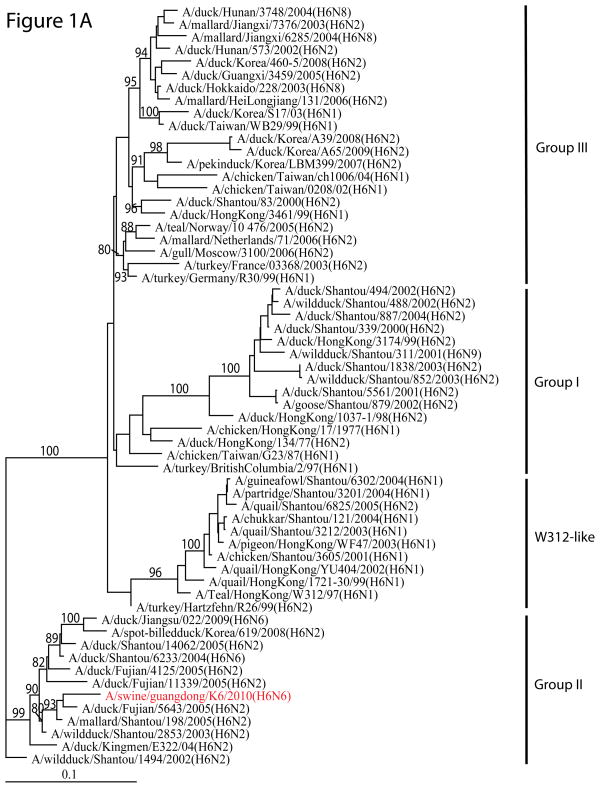
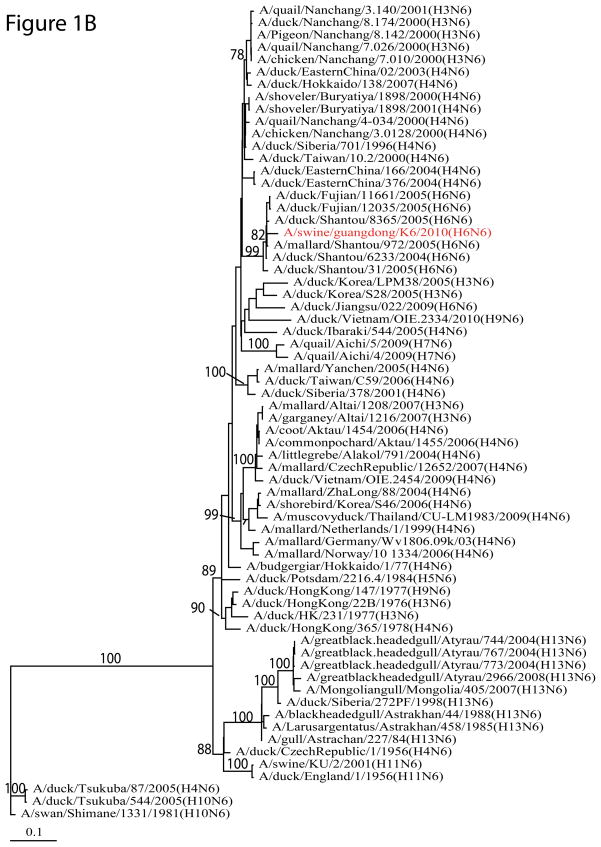
Phylogenetic analysis showed that H6N6 SIV originated from domestic aquatic birds. The HA of this H6N6 SIV belongs to the Group II lineage of H6 AIVs circulating in southern China (Figure 1A). The NA, PB1, PA, NP, NS, and MP from H6 AIVs were also from waterfowl (Figure 1B and Supplementary Figure 1). The PB2 gene was phylogenetically close to those in H5N1 highly pathogenic avian influenza viruses (HPAIVs). The nucleotide sequence identity between each segment of A/swine/Guangdong/K6/2010 (H6N6) and its potential progenitor identified in the public database varied from 92.5% to 98.8%.
Figure 1. Phylogenetic analysis of H6N6 swine influenza virus isolated from southern China.
(A) The phylogenetic tree for HA gene; (B) The phylogenetic tree for NA gene. The phylogenetic trees were constructed using maximum likelihood implemented in GARLI version 0.96, and the bootstrap values were generated using neighborhood joining methods implemented in PAUP* with 1,000 replications.
The protein sequence analysis on the HA gene of A/swine/Guangdong/K6/2010 (H6N6) showed that this virus is lacking polybasic amino acids in the connecting peptide. The receptor binding sites, including 130-loop, 190-helix, and 220-loop, of A/swine/Guangdong/K6/2010(H6N6) were similar to those in its progenitor H6 AIVs. However, compared with those in its progenitor, there were two mutations, A222V and G228S, in A/swine/Guangdong/K6/2010(H6N6). A G228S mutation of H5N1 influenza virus was shown to expand the viral binding profile from ciliated cells to both non-ciliated cells and ciliated cells (Ayora-Talavera et al., 2009). Both H2 and H3 human viruses have the same substitutions, Q226L and G228S, whereas those with Q-226 and G-228 (avian and equine viruses) recognize SAa2, 3Gal (Connor et al., 1994). However, further experiments will be required to study whether A222V and G228S change the cell and host tropisms of this H6N6 SIV.
In order to detect the prevalence of such H6N6 in swine population, a total number of 475 swine serum samples were collected from sick pigs at 11 farms at Guangdong, Guangxi, and Hainan Provinces, China. These sera were tested against H6 subtype using hemagglutination inhibition assay, and all sera with a titer equal or more than 4log2 were defined as positive. Our results demonstrated that 16 of these samples (3.4%) were positive against H6 SIV. These positive samples were collected from four swine farms located at the suburb areas in Shenzhen and Yangjiang of Guandong province, and Guining and Guigang of Guangxi province.
In the past several decades, many influenza epidemics and pandemic strains were reported to emerge from southern China. Both 1957 and 1968 pandemic influenza emerged from this area. The precursor of the ongoing H5N1 HPAIVs was identified also in Sanshui, Foshan, a rural area in southern China (Wan, 1998). Since then, various genotypes of these H5N1 viruses have been identified in this area. Since 2007, avian origin H3N2 canine influenza viruses have been identified also in the dogs in southern China (Li et al., 2010). The report of this H6N6 SIV in southern China further confirmed this area is critical for emergence of novel influenza A viruses.
Identification of H6N6 SIV in the domestic swine suggested that the epidemic of H6 SIV has been expanding from resident birds to the swine population. Our serological surveillance demonstrated a 3.4% of H6 infection rate in this area. Due to the potential of H6 influenza A viruses causing human infection, it is possible that more transmissible H6 viruses may be generated in swine through mutations and reassortments. Continuous monitoring of this virus in swine population is needed.
Supplementary Material
Acknowledgments
We thank Dr. John Harkness for his editorial support. This study was supported by China National High Technology Research and Development Program (China “863” Program 2006AA10A204), Guangdong agricultural class projects (No. 2007A020300006-4) to G. Zhang. L. Long and X.-F. Wan was supported by NIH NIAID RC1AI086830 and NSF-EPS-0903787 subaward 012156-007 to X.-F. Wan. The content is solely the responsibility of the authors and does not represent necessarily the official views of the National Science Foundation, the National Institute Of Allergy And Infectious Diseases, or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abolnik C, Bisschop S, Gerdes T, Olivier A, Horner R. Outbreaks of avian influenza H6N2 viruses in chickens arose by a reassortment of H6N8 and H9N2 ostrich viruses. Virus Genes. 2007;34:37–45. doi: 10.1007/s11262-006-0007-6. [DOI] [PubMed] [Google Scholar]
- Ayora-Talavera G, Shelton H, Scull MA, Ren J, Jones IM, Pickles RJ, Barclay WS. Mutations in H5N1 influenza virus hemagglutinin that confer binding to human tracheal airway epithelium. PLoS One. 2009;4:e7836. doi: 10.1371/journal.pone.0007836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare AS, Webster RG. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- Cheung CL, Vijaykrishna D, Smith GJ, Fan XH, Zhang JX, Bahl J, Duan L, Huang K, Tai H, Wang J, Poon LL, Peiris JS, Chen H, Guan Y. Establishment of influenza A virus (H6N1) in minor poultry species in southern China. J Virol. 2007;81:10402–10412. doi: 10.1128/JVI.01157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin PS, Hoffmann E, Webby R, Webster RG, Guan Y, Peiris M, Shortridge KF. Molecular evolution of H6 influenza viruses from poultry in Southeastern China: prevalence of H6N1 influenza viruses possessing seven A/Hong Kong/156/97 (H5N1)-like genes in poultry. J Virol. 2002;76:507–516. doi: 10.1128/JVI.76.2.507-516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- Huang K, Bahl J, Fan XH, Vijaykrishna D, Cheung CL, Webby RJ, Webster RG, Chen H, Smith GJ, Peiris JS, Guan Y. Establishment of an H6N2 influenza virus lineage in domestic ducks in southern China. J Virol. 2010;84:6978–6986. doi: 10.1128/JVI.00256-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Chang PC, Shien JH, Cheng MC, Chen CL, Shieh HK. Genetic and pathogenic characterization of H6N1 avian influenza viruses isolated in Taiwan between 1972 and 2005. Avian Dis. 2006;50:561–571. doi: 10.1637/7640-050106R.1. [DOI] [PubMed] [Google Scholar]
- Li S, Shi Z, Jiao P, Zhang G, Zhong Z, Tian W, Long LP, Cai Z, Zhu X, Liao M, Wan XF. Avian-origin H3N2 canine influenza A viruses in Southern China. Infect Genet Evol. 2010;10:1286–1288. doi: 10.1016/j.meegid.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WE, Schutten M, Olsen B, Osterhaus AD, Fouchier RA. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 2007;3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KP, Setterquist SF, Capuano AW, Gray GC. Infection due to 3 avian influenza subtypes in United States veterinarians. Clin Infect Dis. 2007;45:4–9. doi: 10.1086/518579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C. Pigs as “mixing vessels” for the creation of new pandemic influenza A viruses. Med Principles Practice. 1990;2:65–71. [Google Scholar]
- Shortridge KF, Stuart-Harris CH. An influenza epicentre? Lancet. 1982;2:812–813. doi: 10.1016/s0140-6736(82)92693-9. [DOI] [PubMed] [Google Scholar]
- Wan X-F. College of Veterinary Medicine. South China Agricultural University; Guangzhou: 1998. Isolation and characterization of avian influenza viruses in China. [Google Scholar]
- Woolcock PR, Suarez DL, Kuney D. Low-pathogenicity avian influenza virus (H6N2) in chickens in California, 2000–02. Avian Dis. 2003;47:872–881. doi: 10.1637/0005-2086-47.s3.872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.