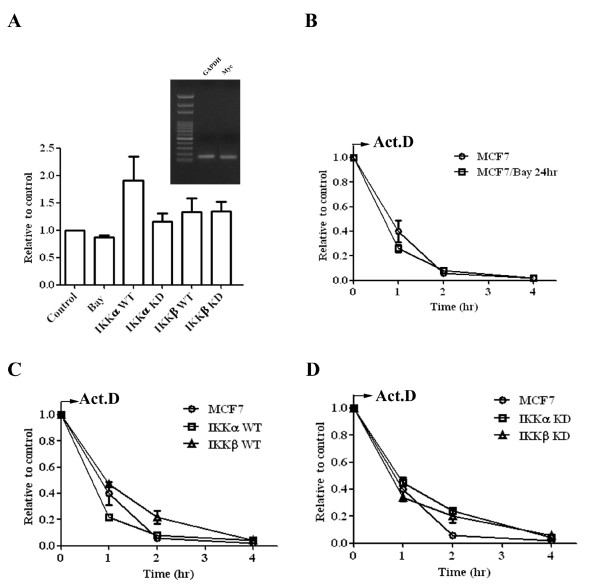
Figure 3.
IKKα and IKKβ increase Myc in a transcription-independent manner. (A) MCF7 cells were treated with 10 μM Bay11-0782 for 12 hours. The RNA was extracted from indicated cells and random-primed reverse transcribed into cDNA. The relative expression of Myc was determined by qPCR. The expression of GAPDH was used as an internal control. The specificity of the primer set for Myc and GAPDH was demonstrated by agarose electrophoresis of PCR product (inserted figure) and by analysis of dissociation curve (data not shown). Each data represents mean ± SD calculated from two independent experiments. To determine the degradation rate of Myc mRNA, the cDNAs were prepared from (B) MCF7 cells were treated with 10 μM Bay11-0782 for 12 hours, (C) MCF7, wild-type IKKα and IKKβ transfected cells, and (D) MCF7, kinase-dead IKKα and IKKβ transfected cells. The relative level of Myc mRNA was determined by qPCR and expressed along a time course after adding actinimycin D. Each data represents mean ± SD calculated from two independent experiments.