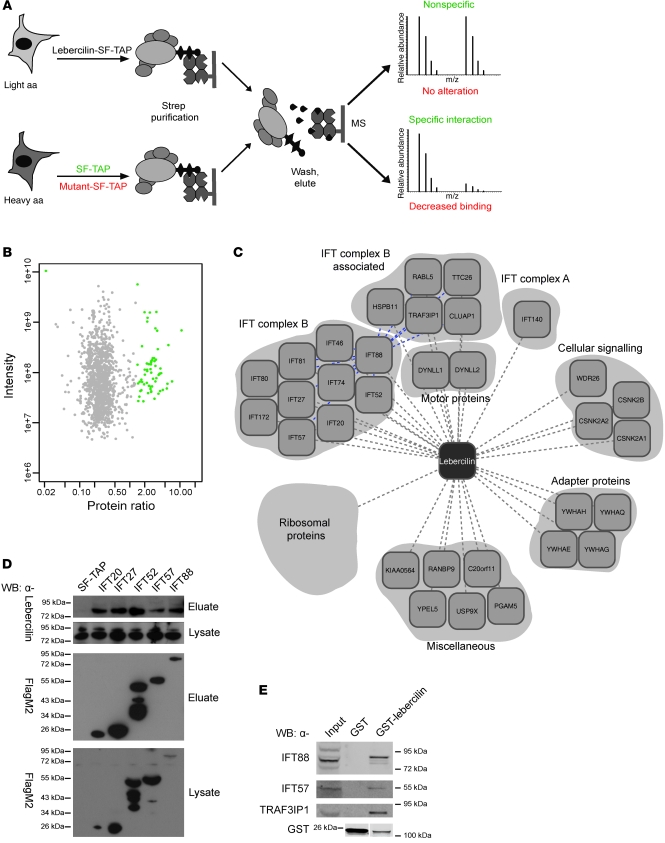
Figure 1. Quantitative protein complex analysis of lebercilin.
(A) SILAC/AP approach to detect specific protein complex components. Identification of significantly enriched proteins compared with control (SF-TAP; green), and analysis of the effect of 2 LCA-causative mutations in lebercilin (p.P493TfsX1 and p.Q279X) on protein complex formation, by comparing the lebercilin complex with complexes formed by the mutants (red) in SILAC-labeled HEK293T cells. MS, mass spectrometry. (B) Detection of specific protein complex components. Plotted are log10 ratios and log10 intensities for each protein identified and quantified in at least 2 of 3 biological replicates. Significantly enriched proteins (P < 0.001) are plotted in green. (C) Protein complex of wild-type lebercilin. Gene names for each protein are shown (see Supplemental Table 1). Blue lines denote interconnectivity, as determined by IFT88 SF-TAP analysis (Supplemental Table 3). (D) Confirmation of IFT-lebercilin association by detecting endogenous lebercilin in eluates of IFT20, IFT27, IFT52, IFT57, and IFT88 SF-TAP experiments. Shown are the lysates as control for lebercilin input. All SF-TAP tagged constructs were expressed and purified, as demonstrated by Western blot using anti-FlagM2. Lebercilin was detected by anti-lebercilin (SN2135) in all eluates, but not in the SF-TAP control, confirming the SILAC/AP results. (E) Confirmation of the association of IFT and IFT-associated proteins with lebercilin in retina by detecting endogenous IFT88, IFT57, and TRAF3IP1 in a GST pulldown of lebercilin, but not in the GST control. Both GST-lebercilin and GST alone were expressed. Lanes for the anti-GST blot were run on the same gel but were noncontiguous (white line).