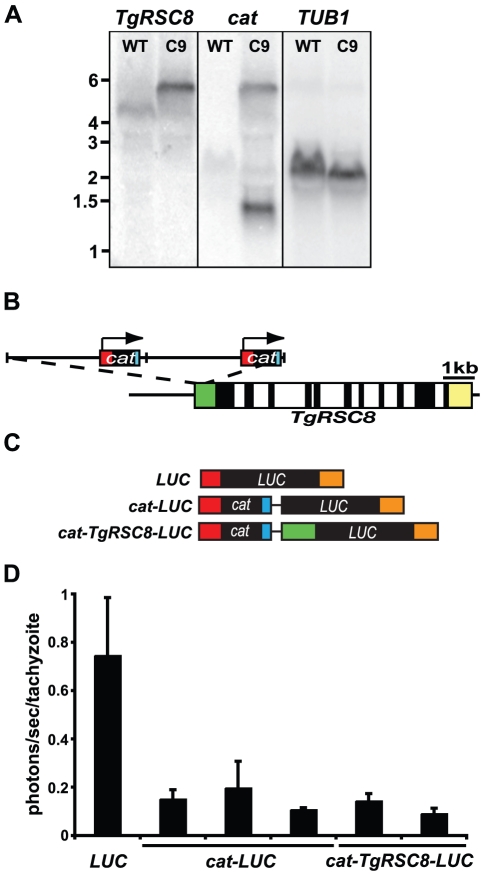
Figure 2. Characterization of the TgRSC8 locus and product.
A. Northern hybridization indicates TgRSC8 is affected in C9. Total RNA from PruΔHPT (WT) or mutant C9 was probed with fragments of TgRSC8 and cat. TUB1 signal was used to assess equivalence of loading. B. Map of the insertion site in strain C9. The TgRSC8 locus is shown with exons (black), introns (white), and 5′ and 3′ UTRs (green and yellow, respectively) to scale. Insertion of 2 copies of pT230-TUB5/CAT (separated by vertical hatch marks) occurred in the 5′ UTR of TgRSC8. The cat gene and UTRs (red) are indicated on the plasmid cartoon, including the direction of transcription from the TUB1 promoter (arrow). C. Transcripts from luciferase reporter constructs. Maps of possible full-length transcripts from constructs containing a monocistronic LUC gene (LUC), polycistronic LUC encoded downstream of cat (cat-LUC), and polycistronic LUC with a partial TgRSC8 5′ UTR (green) between cat and LUC (cat-TgRSC8-LUC) are shown to scale. Additional noncoding sequences, including the TUB1 5′ and 3′ UTR (red and orange, respectively), and SAG1 3′ UTR (blue) are indicated. Vector sequences are shown as black lines. D. Luciferase reporters indicate translation occurs at the downstream locus of a polycistronic transcript in T. gondii. Activity is shown for reporter construct transformants whose transcripts are diagrammed in C, and is displayed as photons per second per tachyzoite. Shown is the average and standard deviations of three independent experiments.