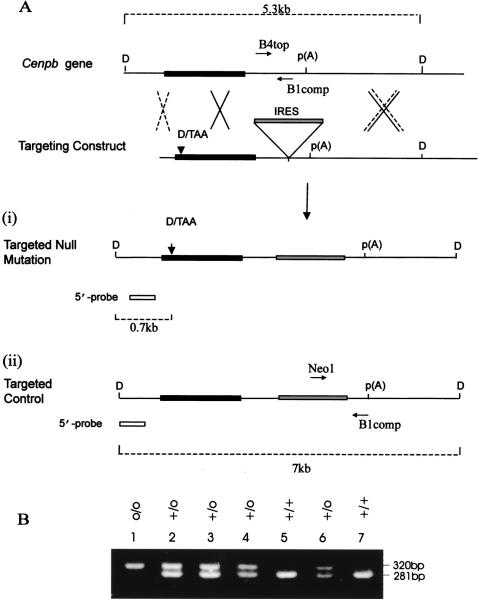
Figure 1.
Strategies for production and screening of Cenpb disrupted and targeted control mice. (A) Intronless wild-type Cenpb gene showing the 1.8-kb coding region (solid box) and polyadenylation p(A) site. Recombination at the sites shown by the dotted crosses results in the incorporation of the targeting construct containing a translational frameshift oligonucleotide (D/TAA) at the 5′ coding region and an IRES-selectable marker cassette (shaded box) within the 3′-untranslated region (i). This recombination event results in the disruption of the Cenpb gene. An alternative recombination event at sites indicated by the solid crosses, one of which is 3′ of the D/TAA frameshift mutation, results in the introduction of the IRES-selectable marker cassette but not the frameshift mutation (ii). This targeted allele served as a control for any positional effect the inserted IRES-selectable marker cassette may have on the phenotype of the mice. The Cenpb wild-type gene (+), null allele (−), and targeted control allele (o) allele were detected as described previously (Hudson et al. 1998) as 5.3-, 0.7-, and 7-kb bands (broken lines), respectively, by DraI (D) digestion and Southern blot hybridization using a 5′-probe (open box) (see Hudson et al. 1998). Once the heterozygous ES cell lines carrying the + and o alleles were identified, they were used for the production of the targeted control (o/o) mice. Subsequent genotype screening for these mice was based on PCR analysis using primers B4top and B1comp, which gave a 281-bp product for the + allele, and primers Neo1 and B1comp which gave a 320-bp product for the o allele. (B) PCR analysis of mouse progeny from a +/o × +/o cross using a combination of the primers B4top, B1comp, and Neo1, showing the +/+ (lanes 5,7), +/o (lanes 2,3,4,6), and o/o (lane 1) genotypes.