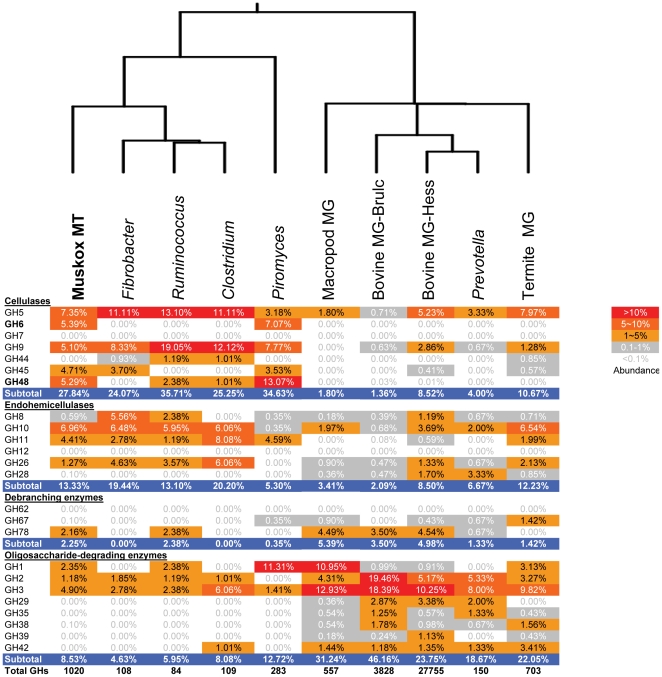
Figure 4. Comparison of the carbohydrate active enzymes identified from muskoxen rumen metatranscriptome (using all assembled contigs) with those of four other foregut metagenomes and five rumen/anaerobic microorganisms.
The percentages of each enzyme family were shown in the cells. Refer to Table S4 for a complete comparison. Dendrogram on the top indicates the relationship of the GHs identified based on similar percentage distribution. Muskox MT: Muskoxen rumen metatranscriptome; Fibrobacter: Genome of Fibrobacter succinogenes S85; Ruminococcus: Genome of Ruminococcus flavefaciens; Clostridium: Genome of Clostridium thermocellum; Piromyces: EST sequence of Piromyces sp. E2; Macropod MG: Macropod foregut microbiome [11]; Termite MG: Termite hindgut microbiome [13]; Bovine MG-Hess: Bovine Rumen microbiome by Hess et al [12]; Bovine MG-Brulc: Bovine Rumen microbiome by Brulc et al [10]; and Prevotella: Genome of Prevotella ruminicola.