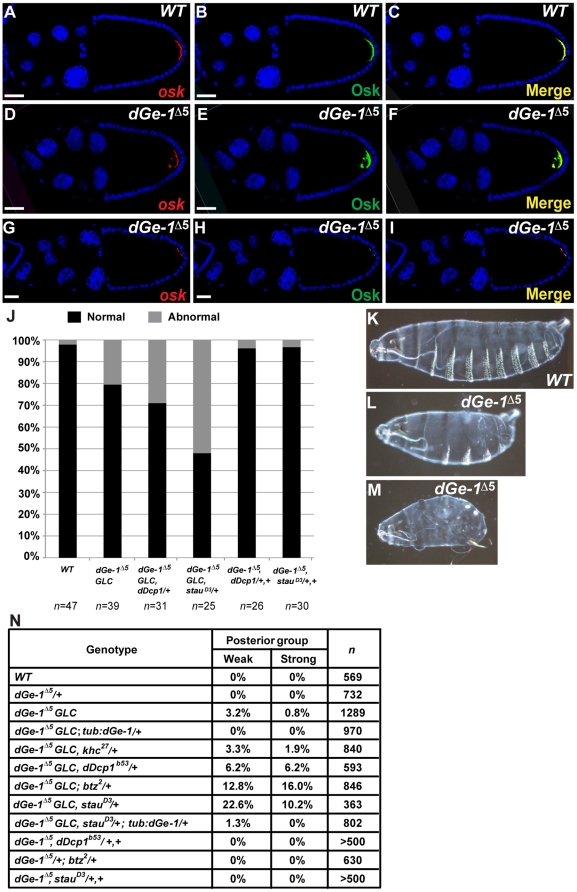
Figure 3. dGe-1 cooperates with Stau and dDcp1 in osk mRNA localization and embryonic patterning.
(A–I) Simultaneous detection of osk mRNA (red) and protein (green) by FISH coupled with immunodetection using a rabbit anti-Osk antibody. osk mRNA (A) and protein (B) colocalize in a posterior crescent at the posterior pole of wt oocytes at S10. (D–I) Examples of the aberrant distribution of osk mRNA and protein in dGe-1Δ5 GLC egg-chambers. Overlays of osk mRNA and Osk protein signals (C, F, I). DNA stained with 4,6-diamidino-2-phenylindole (DAPI) (blue). (J) Quantification (%) of osk mRNA localization patterns in S10 egg-chambers in different genetic backgrounds. Normal and abnormal osk mRNA localization are represented as black and grey bars, respectively. n represents the number of embryos analyzed. (K–M) Loss of maternal dGe-1 causes aberrant cuticle patterning in some embryos derived from dGe-1Δ5 GLC. Lateral view of embryos oriented anterior to the left, ventral side down. (K) In a wt embryo, posterior structures represented by the abdominal denticle belts are clearly visible along the ventral side. In a portion of embryos derived from dGe-1Δ5 GLC, several denticle belts are missing (L) and, in extreme cases, all are absent (M). (N) Quantification of posterior group phenotypes produced in different genetic backgrounds. Embryos lacking at least one (weak) or all (strong) abdominal denticle belts were quantified. n represents the number of embryos analyzed. Bar, 50 µm.