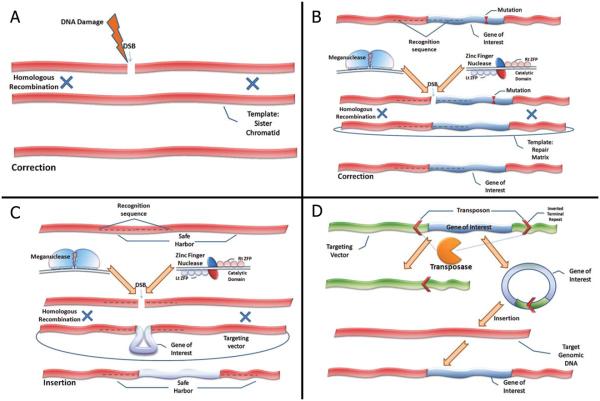
Figure 1. Novel mechanisms for gene correction and insertion.
(A) Homologous recombination (HR) is an important cellular repair mechanism of DNA double-strand breaks (DSB) that occur spontaneously or following exposure to reactive oxidase species, alkylating agents or radiation. Correction is achieved by recombination with the homologous sequence coded on the sister chromatid or a homologous chromosome. (B) Both Homing Endonucleases (HEs) and Zinc Finger Nucleases (ZFN) have the ability to induce DSBs at specific DNA target sequences. HEs and ZFNs may be engineered to recognize genomic regions that are close to disease-causing mutations. ZFN are heterodimers produced by the artificial coupling of zinc finger proteins (in the figure: left ZFP, Lt ZFP; right ZFP, Rt ZFP) that bind to the specific DNA target and a DNA cleavage domain. HEs (also known as meganucleases) are heterodimers produced by modifications of naturally occurring enzymes such as I-SceI. When used in combination with a repair matrix homologous to the DNA target area (but containing the normal sequence), they may promote HR and correction of the mutation. (C) HEs and ZFNs may also be engineered to recognize locations of the genome (“safe harbors”) that do not contain oncogenes or genes encoding for micro-RNAs. When used in combination with a suitable repair matrix, containing the gene of interest flanked by 5′ and 3′ homology regions to the target DNA sequence, these HEs and ZFNs may allow for insertion of a functional gene copy into the “safe harbor”. (D) An alternative approach utilizes transposons which are DNA elements that spontaneously translocate from a specific chromosomal location to another. The transpoase enzyme recognizes inverted terminal repeats (ITR) and cleaves them, thus producing the transposon, which is later re-integrated into a different locus. Introduction of a matrix that codes for the gene of interest, flanked by the ITRs will result in excision of the gene and its re-integration into a genomic locus.