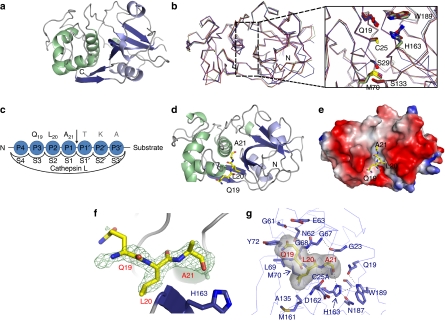
Figure 1. Crystal structures of apo-mC25A and the mC25A and histone H319−33 peptide complex.
(a) The crystal structure of the apo form of the mature, inactive cathepsin L mutant, mC25A. (b) Alignment of the apo-C25A structure with other cathepsin structures of PDB codes 1NPZ, 1VSN and 3BC3 reveals no perturbation in global structure or on the conformation of residues comprising the active-site cleft on mutation of the catalytic cysteine. (c) The standard nomenclature designating the peptide-binding subsites of the cathepsin L active-site-binding cleft. The substrate residues apparent in the crystal structure are indicated in bold. (d) The crystal structure of the mC25A cathepsin L mutant in complex with a peptide derived from the human H3 tail corresponding to residues 19–33 of histone H3 (QLATKAARKSAPATG). Only residues Q19, L20 and A21 could be placed. (e) The Q19–A21 segment of the substrate peptide was fitted into unprimed subsites. The electrostatic surface of mC25A is shown with regions of negative charge indicated in red and positive charges in blue. (f) The Fo–Fc difference density at 3σ contour. The placement of L20 in the S2 subsite was unambiguous. (g) The H3L20 residue occupies the S2 subsite, where it makes a variety of van der Waals contacts with the indicated mC25A side chains. As predicted from biological data, H3A21 occupies the S1 subsite where the S1–S1′ peptide bond would be oriented for nucleophilic cleavage.