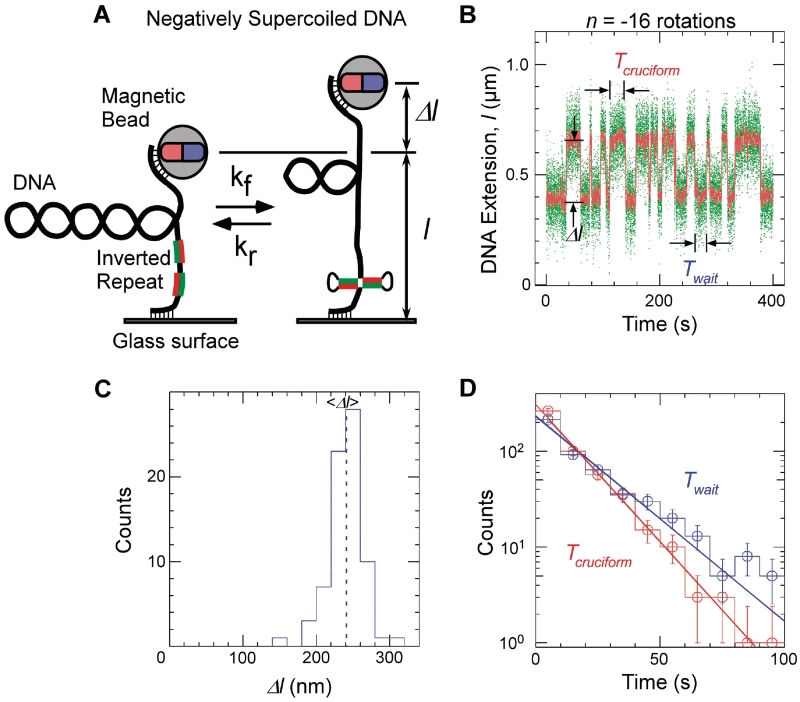
Figure 1.
Single-molecule detection of cruciform extrusion. (A) Sketch of the experiment. A single DNA molecule containing an inverted repeat is tethered at one end to a glass surface and at the other end to a magnetic bead. A pair of magnets serves to control the vertical extending force, F, applied to the DNA and the number of rotations, n, imposed on the bead and hence the DNA supercoiling. The DNA end-to-end extension, l, is determined by measuring the position of the bead above the surface in real-time by computer-aided videomicroscopy. When the inverted repeat extrudes into a cruciform, unwinding of B-DNA titrates out negative supercoils and an increase, Δl, in DNA extension is observed. (B) Real-time data showing abrupt, reversible, changes in extension for Charomid 9-5 kb DNA (F ∼ 0.45 pN, n = −16 turns). Green points correspond to real-time data acquisition (60 Hz) while red points correspond to raw signal averaged over ∼1 s. Both structural (Δl) and kinetic (Twait and Tcruciform) features of the reaction can be obtained from these time-traces. (C) Histogram of transition amplitudes. The dashed line indicates the mean <Δl> = 241 ± 3 nm (n = 74 points). (D) Histograms of Twait (blue) and Tcruciform (red) and single-exponential fits to data. Mean lifetimes are <Twait> = 20.2 ± 1.2 s (n = 490 events) and <Tcruciform> = 13.5 ± 0.9 s (n = 489 events).