Figure 2.
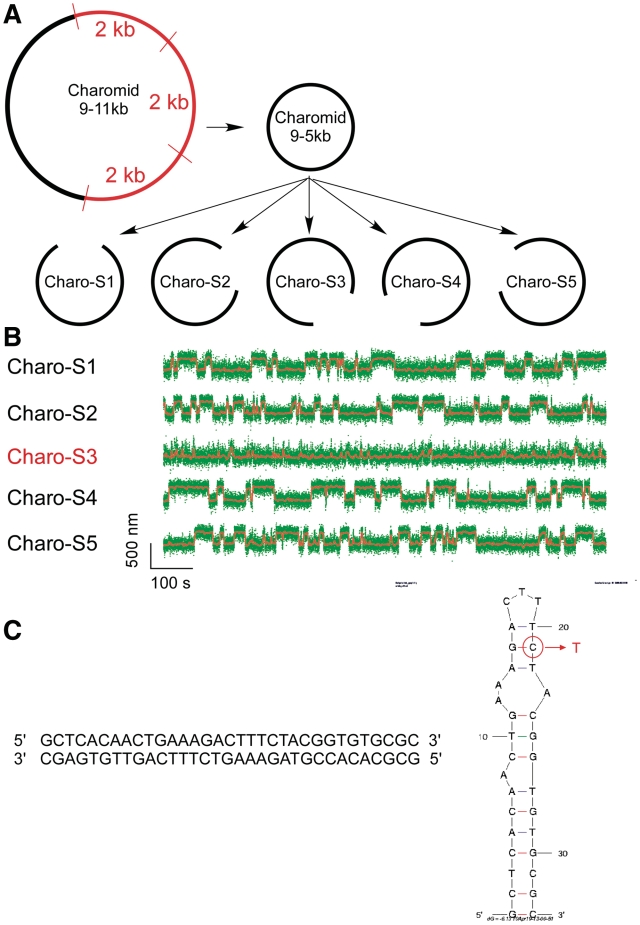
Single-molecule screen of Charomid 9-5 kb is used to identify the Charomid X quasipalindrome. (A) Screen strategy. Charomid 9-5 kb is derived from Charomid 9-11 kb by removing the 6 kb region of the plasmid, corresponding to three consecutive repeats of a 2-kb fragment (35). Then, five constructs are derived from Charomid 9-5 kb using the PCR reaction, each construct lacking a different 1 kb segment of the plasmid. (B) Time-traces for the five constructs described above: first level of screening. DNA constructs were unwound by n = −12 turns using the magnetic trap, and their extension monitored in real-time. The Charomid 9-5 kb displays the same behavior as the Charomid 9-11 kb plasmid, indicating that the repeat region is not responsible for the observed fluctuations. The Charo-S3 construct is the only one of the five constructs to lack the canonical fluctuations observed on the original plasmid, indicating that the sequence responsible for this behavior is located in the corresponding 1 kb of DNA, which is specifically lacking in the construct. Fast residual fluctuations indicate denaturation or cruciform extrusion on secondary sequences in the plasmid (C) Screen results. The above process was reiterated two times to narrow down the sequence responsible for the observed fluctuations. The 33-bp Charomid X sequence responsible for the observed fluctuations is a quasipalindrome, which extrudes to form a complex and unstable cruciform. A putative minimum energy fold derived from mfold for high-salt conditions (0.5 M NaCl) is presented; in the lower salt conditions initially used here, the 3-bp separating the two apical loops are unlikely to be stable at room temperature, implying that the apical loop is in fact the sum of the two smaller loops. This is consistent with the extensive amount of unwinding estimated to be present in the transition state. The mutation made to increase the loop size involved converting the G circled in red into a T, disrupting a crucial base pair for high-salt conditions but not low-salt conditions (Table 1).