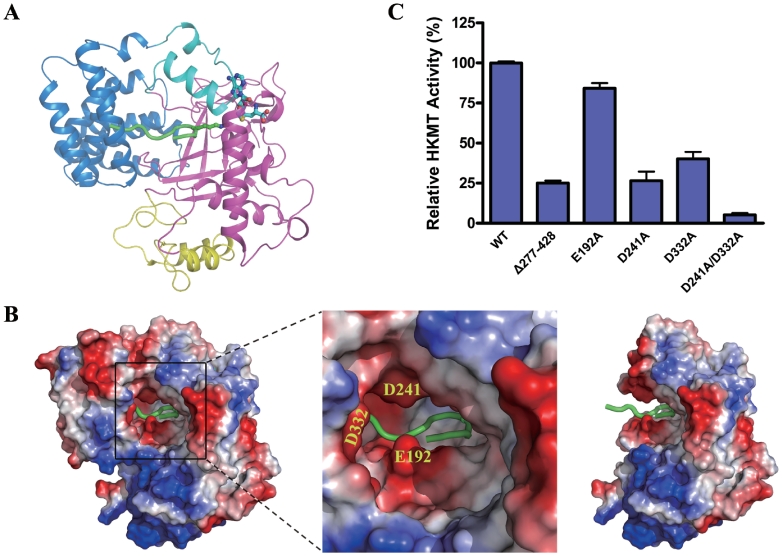
Figure 2.
Histone binding pocket. (A) Potential histone peptide binding site. Superposition of the Smyd3 and SET7/9 (PDB code 1O9S) structures was performed as in Figure 1B. Smyd3 is shown in a ribbon representation with the same color coding as in Figure 1A. For simplicity, for SET7/9 only the bound histone peptide is shown in a ribbon representation and colored in green and the side chain of the methyllysine of the histone peptide is shown with a ball-and-stick model and colored accordingly. The cofactor is also shown with a ball-and-stick model. (B) Electrostatic potential surface of the potential histone peptide binding pocket in Smyd3. The surface charge distribution is displayed as blue for positive, red for negative, and white for neutral. A close-up view of the pocket (middle panel) shows that several acidic patches are present at the opening of the binding pocket. Some of the acidic residues in these patches are labeled. The TPR domain forms part of the substrate binding pocket and removal of the TPR domain would leave an incomplete pocket (right panel). (C) HKMT activity assays of the wild-type Smyd3 and the mutants with truncation or mutations at the substrate binding pocket. The activities of the truncate with deletion of the C-terminal region (Δ277–428) and the mutants carrying one or two point mutations of the residues potentially involved in histone binding were determined. Activity is shown as the relative activity of the proteins normalized to that of the wild-type protein. The experiments were performed in triplicates and the error bars indicate the standard deviation.