Abstract
While searching for compounds with antimalarial activity, two new naphthoquinones, delitzchianones A (1) and B (2), were separated from Delitzchia winteri, an endophytic fungus from Costa Rica. The same search also led to a new 8-acetoxy pestalopyrone (3) and the known compound, pestalopyrone (4) from another Costa Rican endophytic fungus, Phomatospora bellaminuta. The structures of the three new compounds 1, 2 and 3 were established with extensive NMR and MS analyses. All four compounds were tested for activity in a growth / no growth Dd2 assay, but only compound 4 had measurable activity with an IC50 value of 37 μM.
Keywords: endophytic fungi, anti-malaria, naphthoquinones, δ-lactones
While malaria poses a major public health burden – 300 to 500 million cases a year – to populations in tropical and subtropical areas around the world, the most affected areas are in southern Africa where the majority of the 1 to 1.5 million annual deaths occur. Four malaria parasite species affect humans, but Plasmodium falciparum is both the most widespread and most lethal. Four different compound classes have provided drugs that are currently useful for malaria, but the development of parasite strains resistant to all commonly used drugs has eroded our ability to combat the disease.1
In a continuing search for antimalarial natural products, endophytic fungi from Costa Rica, a biodiversity hot spot,2 have been examined. This report deals with two extracts (CR237A and CR1092F), which were obtained from the endophytic fungi Delitzchia winteri and Phomatospora bellaminuta, respectivly.3 Neither species had been examined previously, although flutimide, (Z)-5-(2-methylpropyl)-3-(2-methylpropylidene)-1-hydroxy-3H-pyrazine-2,6-dione, which has been shown to selectively inhibit cap-dependent endonuclease activity of influenza virus A, was isolated from Delitschia cofertaspora.4
CR237A was first fractionated over a C18 SPE column, and fraction III was further separated by C-18 prep-HPLC to yield compounds 1 and 2; Compounds 3 and 4 were obtained from CR1092F after solvent partition and HPLC purification.5 Compound 4 was identified as pestalopyrone.6
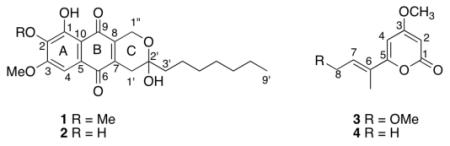
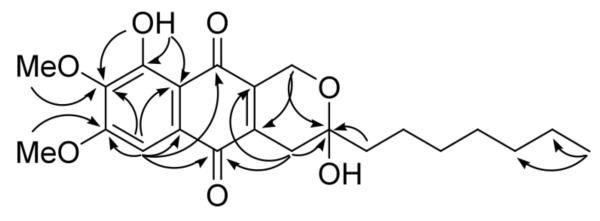
Compound 17 was isolated as yellow powder. The HRMS (positive-ion mode) had an ion peak at m/z 387.1819, consistent with a molecular composition of C22H27O6 ([M-H2O+H]), calcd 387.1808). A molecular formula of C22H28O7 requires nine double-bond equivalents. The 1H NMR spectrum of 1 in CDCl3 showed one chelated hydroxyl, one aromatic proton, two methoxys, eight methylenes, and one methyl group (Table 1). Its 13C NMR spectrum exhibited twenty-two signals, including two carbonyls, eight sp2 carbons, one oxygenated sp3 quaternary carbon, two methoxys, and one methyl group. These were further confirmed by the HSQC. In the HMBC spectrum (Figure 1), the aromatic proton (H-4, δH 7.27 s) showed correlations to C-2 (δC 141.4), C-3 (δC 157.9), C-5 (δC 127.6), C-6 (δC 182.5), C-9 (δC 187.5, a weak 5J correlation with a W shape), and C-10 (δC 110.7). On the other hand, the chelated hydoxyl proton (δH 12.05 s) correlated to C-1 (δC 155.7), C-2, and C-10, indicating that this chelated hydoxyl group (1-OH) was para to the aromatic proton (H-4). Also on the same ring (ring A), 2-OMe (δH 4.05 s) and 3-OMe (δH 3.99 s) exhibited HMBC correlations to C-2 and C-3, respectively. The methylene at δH 4.68/4.73 (d, J = 18.6 Hz, H2-1″) showed correlations to C-7 (δC 141.3) and C-2′ (δC 96.0), and another methylene at δ 2.79/2.53 (d, J = 20.1 Hz, H2-1′) correlated to C-2′, C-8 (δH 140.4) and C-6. From the above information, it could be deduced that ring C was a 2,3-dihydropyran and the oxygenated methylene (H2-1″) was at the same side as the chelated hydroxyl group (1-OH). The low field chemical shift of the quaternary sp3 carbon C-2′ (δ 96.0) indicated that it must be a hemiacetal connected to a hydroxyl group and a heptyl, –(CH2)6CH3. Hence, the structure of 1 was determined as shown.
Table 1.
1H and 13C NMR Data of Compounds 1 and 2
| # | 1 (1H)a | 2 (1H)a | 2 (1H)b | # | 1 (13C)c | 2 (13C)d |
|---|---|---|---|---|---|---|
| 1 | 1 | 155.7 | 154.7 | |||
| 2 | 2 | 141.4 | 142.7 | |||
| 3 | 3 | 157.9 | 154.4 | |||
| 4 | 7.27 (s) | 7.27 (s) | 7.25/7.27 (s) | 4 | 104.3 | 107.3/107.7 |
| 5 | 5 | 127.6 | 129.5/130.5 | |||
| 6 | 6 | 182.5 | 182.8 | |||
| 7 | 7 | 141.3 | 140.8 | |||
| 8 | 8 | 140.4 | 140.8 | |||
| 9 | 9 | 187.5 | 189.9 | |||
| 10 | 10 | 110.7 | 111.9 | |||
| 1′ | 2.79 (d 20.1) | 2.78 (d 19.8) | 2.63/2.67 (d 19.8) | 1′ | 31.7 | 32.3 |
| 2.53 (d 20.1) | 2.52 (d 19.8) | 2.41/2.43 (d 19.8) | ||||
| 2′ | 2′ | 96.0 | 96.8 | |||
| 3′ | 1.80 (m) | 1.77 (m) | 1.73 (m) | 3′ | 42.2 | 42.8 |
| 4′ | 1.2~1.6 (m) | 1.2~1.6 (m) | 1.2~1.6 (m) | 4′ | 22.6 | 23.7 |
| 5′ | 1.2~1.6 (m) | 1.2~1.6 (m) | 1.2~1.6 (m) | 5′ | 30.6 | 31.0 |
| 6′ | 1.2~1.6 (m) | 1.2~1.6 (m) | 1.2~1.6 (m) | 6′ | 29.7 | 30.8 |
| 7′ | 1.2~1.6 (m) | 1.2~1.6 (m) | 1.2~1.6 (m) | 7′ | 29.2 | 30.4 |
| 8′ | 1.2~1.6 (m) | 1.2~1.6 (m) | 1.2~1.6 (m) | 8′ | 23.0 | 24.5 |
| 9′ | 0.90 (t 6.6) | 0.88 (t 6.6) | 0.91 (t 6.6) | 9′ | 14.1 | 14.4 |
| 1″ | 4.73 (d 18.6) | 4.71 (d 18.6) | 4.59 (m) | 1″ | 56.5 | 56.5 |
| 4.68 (d 18.6) | 4.66 (d 18.6) | |||||
| 1-OH | 12.05 | 11.90 (s) | ||||
| 2-OCH3 | 4.05 (s) | 2-OCH3 | 61.0 | |||
| 3-OCH3 | 3.99 (s) | 4.01 (s) | 3.94/3.90 (s) | 3-OCH3 | 57.4 | 58.1 |
δ (ppm) 500 MHz in CDCl3; multiplicities; J values (Hz) in parentheses.
δ (ppm) 500 MHz in CD3OD; multiplicities; J values (Hz) in parentheses.
δ (ppm) 150 MHz in CDCl3.
δ (ppm) 150 MHz in CD3OD.
Figure 1.
HMBC Correlations of 1
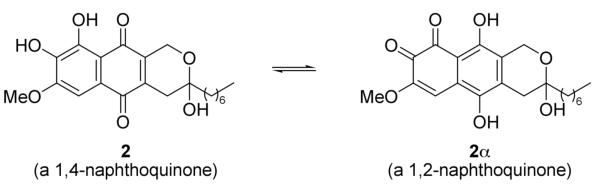
Compound 28 was isolated as red powder, and had a molecular formula of C22H26O6, which is 14 units less than 1. A 1H NMR spectrum in CDCl3 was collected immediately after compound 2 was purified, and only one methoxy at δH 4.01 (s) was observed. The only difference between 1 and 2 was the substituent at the 2-position. Inspection of the 1D and 2D NMR spectra of 2 in MeOH-d4 (Table 1) indicated that the compound consisted of two major tautomers (Figure 2), designated 2 (a para naphthoquinone) and 2αa (an ortho naphthoquinone) in the approximate proportion of 7:6, indicating that compounds 2 was a 2-O-demethyl product of compound 1. Hence, the structure of 2 was determined as shown.
Figure 2.
Tautomerization of 2
Compound 39 was isolated as colorless powder. The NMR data of compound 3 were very similar to those of compound 4 except 8-position and the substituent at C-8. The chemical shifts of H-8 (4.77 d J = 6.0 Hz) and C-8 (60.8) indicated that there was an acetoxy group at 8-position (δH 2.04 s/δC 21.0 and δC 169.5). Hence, compound 3 was determined as 8-acetoxy pestalopyrone.
The two new naphthoquinones, 1 and 2, and the two δ-lactones, 3 and 4, were evaluated in a Plasmodium falciparum (Dd2) assay,10 but only 4 showed marginal activity against Dd2 with an IC50 value of 37 μM.
Supplementary Material
Acknowledgments
This work was generously supported by NIH U01 TW007404 (J.C.) and Medicines for Malaria Ventures (MMV) supported the antimalarial assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Na-Bangchang K, Karbwang J. Fundamental Clin. Pharmacol. 2009;23:387–409. doi: 10.1111/j.1472-8206.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- 2.a Cao S, Ross L, Tamayo G, Clardy J. Org. Lett. 2010 doi: 10.1021/ol101972g. in press (DOI: 10.1021/ol101972g) [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kontnik R, Clardy J. Org. Lett. 2008;10:4149–4151. doi: 10.1021/ol801726k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sequencing and species identification. For identification by internal transcribed spacer (ITS) sequencing, CR237A and CR1092F were cultured in potato dextrose broth for 5 days. The mycelium was then retrieved by filtration and ground to a fine powder in liquid N2. Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega), and large subunit rDNA was amplified by PCR using primers LR5 (5′-TCCTGAGGGAAACTTCG-3′) and LROR (5′-ACCCGCTGAACTTAAGC-3′). PCR products were transformed into E. coli TOP10 cells using a TOPO TA Cloning Kit (Invitrogen), according to manufacturer's protocols. Transformed plasmids were isolated and sequenced at Genewiz (http://www.genewiz.com/). The following consensus sequence were used in a BLAST search against deposited sequences: 237A:CCCCTATGCCCAAATTTGACGATCGATTTGCACGTCAGAACCGCTGCGAGCCTCCACCAGAGTTTCCTCTGGCTTCACCCTATTCAGGCATAGTTCACCATCTTTCGGGTCCCAACAGCTATGCTCTTACTCAAATCCATCCGAAGACATCAGGATCGGTCGATGGTGCGCCAGAGCTCGCGCCCTGGGTCCCACCTCCGTTCACTTTCATTCCGCGCCCGGGCTTGACACCCAAACACTCGCATAGATGTTAGACTCCTTGGTCCGTGTTTCAAGACGGGCCGCTTACGACCATTACGCCAGCATCCTAGCCGAAGCGCGGACCTCAGTCGGGGCTGGCTGCATGACGCCCTGGGCTATAACACTCCCCGAAGAGAGCTACATTCCCAAGGCCTTTCTCCAGCCGCCCCAACTGATGCTGGCCTGCCTGCCGCCGAGTGCACAGGGGACGGACCCCCGATGAACAGCGGCAGCCAAGTCTGGTTGCAAGCGCTTCCCTTTCAACAATTTCACGTGCTGTTTGACTCTCTTTCCAAAGTGCTTTTCATCTTTCGATCACTCTACTTGTGCGCTATCGGTCTCTGGCCAGTATTTAGCTTTAGAAGAAATATACCTCCCATTTAGAGCTGCATTCCCAAACAACTCGACTCGTCGAAGGGGGTTCACATGGCGCAGGCACCTGCCGCGTACGGGGTTCTCACCCTCTCTGACGTCCCGTTCCAAGGAACTTAGACAGGCGNCGTTGCCGAACCACCNTCTGCAAAGTACAACTCGGANCCCGCAAGGAGCCAGATTTCAAATTTGAGCTGTTGCCGCTTCACTCGCCGTTACTGAGGCAAT CR1092F:AGAGGTTGATAGTCTTTCGCCCCCATGCTCATGTTTGACGATCGATTTGCACGTCAGAACCGCTGCGAGCCTCCACCAGAGTTTCCTCTGGCTTCACCCTACACAAGCATAGTTCACCATCTTTCGGGTCCAAGCGGCAAGGCTCTTACTCAAATCCATCCGAAGACTTCAGGATCGGTCGATGGTGCGCCGAGGCTCCCACCTACGTTCACTTTCATTTCGCGTGCGGGTTTTACACCCAAACACTCGCCCTAATGCTTGACTCCTTGGTCCGTGTTTCAAGACGGGTCGCTGGTGACCATTACGCCAGCATCCTTGCAATGCGCGGTCCTCGGTCCCCGCGAGGGCATTGAGCAACGGGCTATAACACTCCCGGAGGAGCCACATTCCCGAGGCCTTTATCCCCCCGCGAGAACCGATGCTGGCCCGAGCCCGGCGGAGTGCACCGGCGAGAACGCCGGATGATCCGCCGGGCGCGAGTCTGGTCACAGGCGCTTCCCTTTCAACAATTTCACGTGCTTTTTAACTCTCTTTTCAAAGTGCTTTTCATCTTTCGATCACTCTACTTGTGCGCTATCGGTCTCTGGCCGGTATTTAGCTTTAGAAGAAATTTACCTCCCGCTTTGAGCAGCATTCCCAAACTACTCGACTCGTCGAAGGAGCTTTACAGAGGCTCGGCGTCCGCCTGTACGGGGCTCTCACCCTCTATGGCGTCCCGTTCCAGGGAACTCGGACGGCGCCTTGCCAAAAGCATCCTCTACAGATTACAACTCGGGCCCTGGGGACCAGATTTCAAATCTGAGCTGTTGCCGCTTCACTCGCCGTTACTGGGGCAATCCCTGTTGGTTTCTTTTCCTCCGCTTATTGATATGGTTAGTTTCAANCGGGGTAA
- 4.Singh SB, Tomassini JE. J. Org. Chem. 2001;66:5504–5516. doi: 10.1021/jo015665d. [DOI] [PubMed] [Google Scholar]
- 5.Culturing and extraction. Agar plugs of CR237A and CR1092F were initially grown at 25°C on yeast malt agar plates supplemented with 30 μg/ml streptomycin and 12 μg/ml chlortetracycline. After one week, agar plugs of this plate were placed in 150 ml of rich seed media in 1 L flasks. They were grown at 25 °C and 150 rpm for 7 days. 450 ml of 0.66% (w/v) malt extract and 10 g HP-20 resin were then added to each flask, and the fungi were cultured under the same conditions for 21 days. The fungal cultures were then held at 25 °C without shaking for 5 days. Extraction of the mycelium was accomplished by three rounds of sonication in ethanol. Rich seed media: 5 g peptone, 10 g dextrose, 3 g yeast extract, 10 g malt extract per 1 L water (pH 6.2). Separation. Extract CR237A was loaded on C-18 SPE and three fractions were collected. Compounds 1 (tR: 33.0 min, 1.5 mg) and 2 (tR: 28.5 min, 1.0 mg) were collected after a C-18 HPLC column (250 × 21.2 mm, 5 μ, 10 ml/min, 80% MeOH for 20 min then to 100% MeOH in 10 min) from fraction 3. Extract CR1092F was was suspended in aqueous MeOH (MeOH-H2O, 9:1, 100 mL) and extracted with hexanes (3 × 100 mL portions). The aqueous layer was then diluted to 70 % MeOH with H2O and extracted with CH2Cl2 (3 × 100 mL portions). After C-18 SPE, the hexanes extract was separated using HPLC to yield compound 3 (tR: 8 min, ~0.1 mg; C-18, Phemonenex, Luna, 250 × 10 mm, 5 μ, 4 ml/min, 45% MeOH) and compound 4 (tR: 9.5 min, 2.0 mg; C-18, Phemonenex, Luna, 250 × 10 mm, 5 μ, 2 ml/min, 60~100% MeOH in 20 min).
- 6.Venkatasubbaiah P, Van Dyke CG, Chilton WS. Phytochemistry. 1991;30:1471–1474. [Google Scholar]
- 7.Delitzchianone A (1): yellow powder; [α]26D +120 (c, 0.06 MeOH); UV (MeOH) λmax 215, 260, 295 (sh), 422 nm; 1H NMR (600 MHz, CD3OD): see Table 1; 13C NMR (150 MHz, CD3OD): see Table 1; HRMS m/z 387.1819 ([M-H2O+H]), calcd for C22H27O6, 387.1808).
- 8.Delitzchianone B (2): red powder; [α]26D +161 (c, 0.08 MeOH); UV (MeOH) λmax 222, 273, 300 (sh), 428 nm; 1H NMR (600 MHz, CDCl3 and CD3OD): see Table 1; 13C NMR (150 MHz, CD3OD): see Table 1; HRMS m/z 373.1719 ([M-H2O+H]), calcd for C21H25O6, 373,1651).
- 9.8-Acetoxy pestalopyrone (3): colorless powder; UV (50% MeOH/H2O) λmax UV (50% MeOH/H2O) λmax 310 nm; 1H NMR (600 MHz, DMSO-d6): δH 1.88 s (H3-9), 2.04 s (OAc), 3.82 s (OMe), 4.77 d J = 6.0 Hz (H2-8), 5.65 br s (H-4), 6.31 br s (H-2), 6.35 t J = 6.0 Hz (H-7); 13C NMR (150 MHz, DMSO-d6): 12.7 (C-9), 21.0 (OCOCH3), 56.8 (OMe), 60.8 (C-8), 89.1 (C-4), 99.3 (C-2), 127.6 (C-6), 127.6 (C-6), 158.1 (C-5), 169.5 (OCOCH3), 170.1 (C-3), 173.8 (C-1); HRMS m/z 239.0925 ([M+H]), calcd for C12H15O5, 239.0920).
- 10.Antimalarial Screen. The antimalarial screen was carried out as previously described.2 Briefly, extracts dissolved in DMSO were arrayed in dilution series in 384-well plates. The solutions were pintransferred to assay plates containing red blood cells parasitized with Plasmodium falciparum. Afluorescent DNA stain (DAPI) was added after a 72-hour incubation period, and the plates imaged to quantify levels of parasitic nuclei.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.