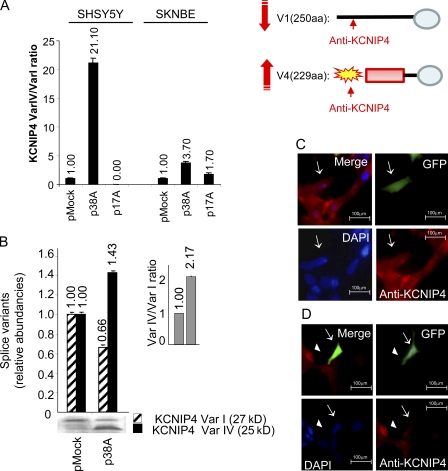
Figure 2.
ncRNA-induced alternative splicing analysis. (A) Real-time RT-PCR detection of KCNIP4 splice variant synthesis in SHSY5Y and/or in SKNBE cells transiently transfected with either 38A or 17A (a noncorrelated PolIII-transcribed ncRNA used as a negative control) expression plasmids. Altogether, these results indicate that each ncRNA specifically influences the splicing of its corresponding protein-coding gene. (B) Western blotting analysis of the 38A-dependent alternative splicing of KCNIP4. In pMock-transfected cells, the relative abundance of the 250-residue-long form of KCNIP4 (Var I, striped bars) is predominant with respect to the alternatively spliced 229-residue-long form (Var IV, shaded bars) that, in turn, is the main KCNIP4 form synthesized by p38A-transfected cells. Quantitative bars and the correspondent error bars (SD) are referred to the mean of three independent experimental determinations (A and B). The p38A-dependent change of Var IV versus Var I protein form ratio is quantitatively reported (inset in B). (C and D) Immunofluorescence detection of KCNIP4 alternative splicing. Antibodies raised against the KCNIP4 N-terminal fragment of the protein form I show a clearly detectable signal in GFP-expressing cells only in the absence of a concomitant 38A overexpression (arrows in C); on the contrary, the signal is absent or very weak in cells transfected with a construct coexpressing 38A and GFP (arrows in D). Untransfected cells, which do not express GFP and 38A, are positive for KCNIP4 (arrowheads in D).