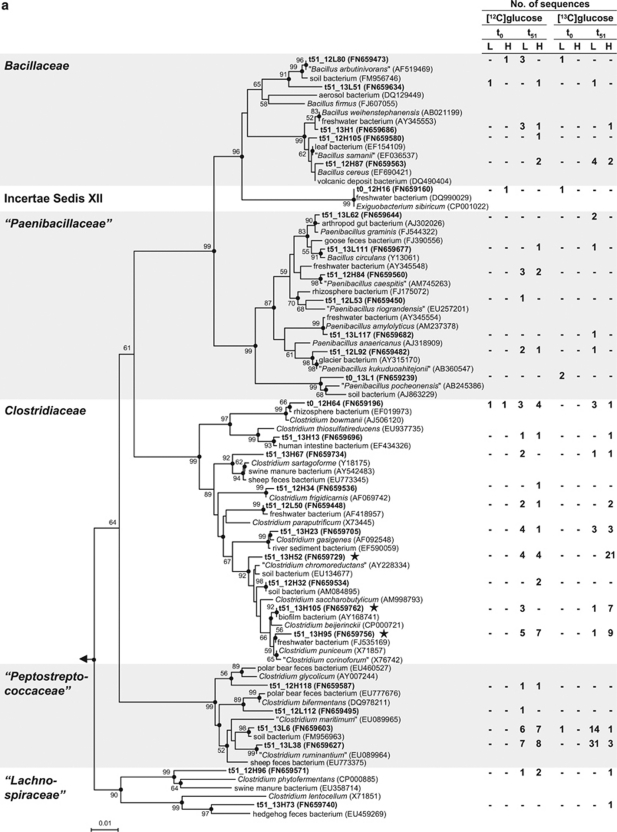
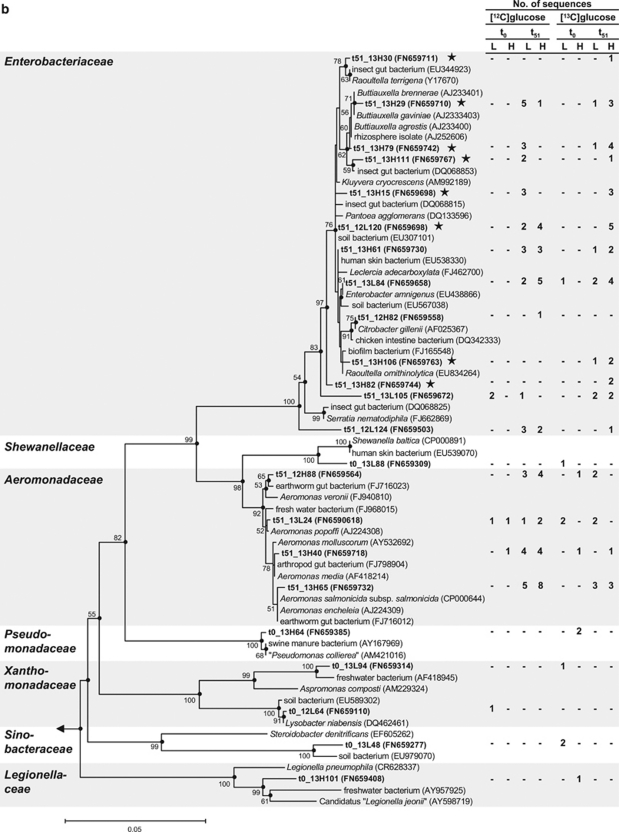
Figure 4.
Phylogenetic tree of 16S rRNA complementaryDNA sequences (bold) retrieved from the earthworm gut and reference sequences of the Firmicutes (a) and Gammaproteobacteria (b). Only representative sequences are shown. The values next to the branches represent the percentages of replicate trees (>50%) in which the associated taxa clustered together in the bootstrap test (10 000 replicates). Dots at nodes indicate confirmation of topology by AxML using the same data set. Labeled taxa are marked with stars. Methanosarcina barkeri (AF028692) was used as outgroup. Accession numbers are in parentheses. Quotation marks indicate nonvalidated taxa (Euzéby, 2010). Abbreviations: t0, at the start of incubation; t51, at 51 h of incubation; L, ‘light' fractions; H, ‘heavy' fractions. (a): The family Veillonellaceae was excluded because the sequence that affiliated with this family (t51_12H90) was shorter than 700 bp. The phylogenetic tree was calculated using the neighbor-joining method (50% minimum similarity filter; 651 valid positions between 104 and 815 of the 16S rRNA gene of E. coli). Bar indicates 0.01 estimated change per nucleotide. (b): The labeled sequence t51_13H112 (next cultivated species Serratia fonticola, AY236502, 99% 16S rRNA gene similarity) was excluded from tree calculation because it was shorter than 700 bp. The phylogenetic tree was calculated using the neighbor-joining method (50% minimum similarity filter; 698 valid positions between 101 and 816 of the 16S rRNA gene of E. coli). Bar indicates 0.05 estimated change per nucleotide.