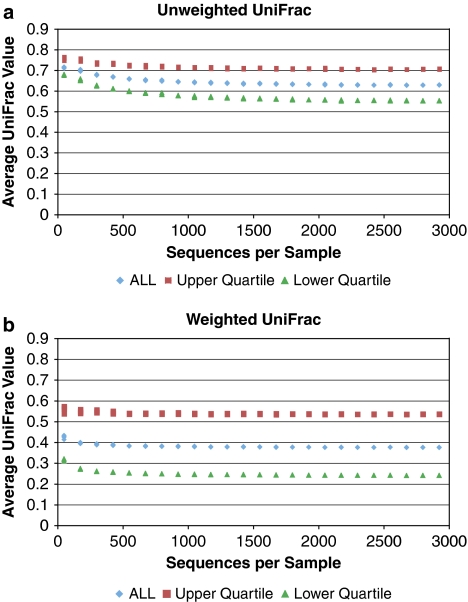
Figure 1.
Rarefaction of data from a study of obese twins (Turnbaugh et al., 2009). This study produced >1 million reads from the V2 region of ribosomal RNA using pyrosequencing. The samples with less than 3000 sequences were first excluded (leaving 112 samples). For five replicate trials, sequences from all 112 samples were subsampled so that each sample had a set number of sequences (between 50 and 2925 with a step size of 125). Pairwise UniFrac values were calculated with both the unweighted (a) and weighted (b) versions for all pairs of samples. To assess the effects of community divergence (the raw UniFrac value) on the sensitivity to sampling, the most similar and most different pairs of samples were identified from the most heavily subsampled data set (2925 sequences per sample) as those in the upper and lower quartile of UniFrac values respectively (calculated separately for unweighted and weighted). The points represent the average UniFrac value at each sample depth for (1) all pairwise comparisons and (2) the pairs that were identified as being in the upper and lower quartiles. Individual points for each of the five replicate trials are plotted, but the values of the replicates were close enough that they are generally on top of each other except for the smallest subsamples.