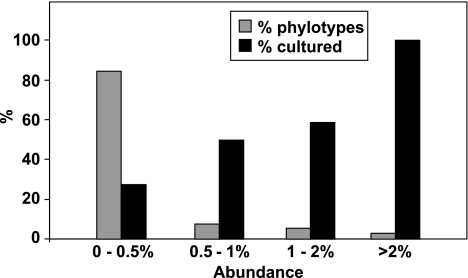
Figure 1.
Culturability of 16S rRNA phylotypes in relation to their abundance. 16S rRNA sequences (5915) obtained from faecal samples of six volunteers (for all four diets) were classified into 320 phylotypes (defined at >98% sequence identity) (Supplementary Table S4). The % of phylotypes showing >98% sequence identity to a cultured bacterium is seen to increase with increasing phylotype abundance. The phylotype frequency distribution is shown as a percentage; actual numbers of phylotypes were 9 (>2% of all sequences), 17 (>1%<2%), 24 (>0.5%<1%) and 270 (<0.5%) (total 320). Data refer to volunteers 16, 20, 22 (diet order RS-NSP) and 19, 23, 24 (diet order NSP-RS).