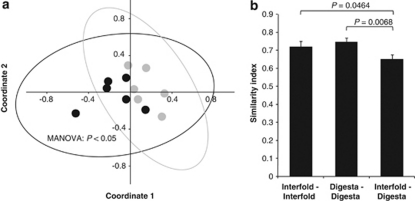
Figure 4.
Lachnospiraceae–Ruminococcaceae populations are distinct in the interfold region compared with the digesta region. Interindividual variation of Lachnospiraceae–Ruminococcaceae was examined by PCR and terminal restriction fragment length polymorphism techniques targeting Lachnospiraceae—Ruminococcaceae-specific 16S rRNA genes. (a) Differences in the structure and composition of this bacterial family between the interfold (black symbols) and digesta (gray symbols) regions were examined by multivariate analysis (n=6). Non-metric multidimensional scaling analysis ordinations derived from the Kulczynski similarity index (presence-absence data). Each symbol is representative of a single sample. Samples are plotted along the first two component axes. The ellipse corresponds to the joint 95% confidence limits. Microbial composition between the two regions was compared using non-parametric MANOVA. (b) Pairwise comparisons of α-diversity (profile similarity between different subjects, that is, interfold versus interfold) and β-diversity (profile similarity across the interfold and digesta regions) were examined by the Kulczynski similarity index (presence-absence data) and inferential statistics. ANOVA and Protected Least-Significant Difference test were used to compare differences in α-diversity and β-diversity. Differences were considered significant at P<0.05.