Abstract
Objective
Genetic association of the IL2/IL21 region at 4q27 has been previously reported in lupus and a number of autoimmune and inflammatory diseases. Herein, using a very large cohort of lupus patients and controls, we localize this genetic effect to the IL21 gene.
Methods
We genotyped 45 tag SNPs across the IL2/IL21 locus in two large independent lupus sample sets. We studied a European-derived set consisting of 4,248 lupus patients and 3,818 healthy controls, and an African-American set of 1,569 patients and 1,893 healthy controls. Imputation in 3,004 WTCCC additional control individuals was also performed. Genetic association between the genotyped markers was determined, and pair-wise conditional analysis was performed to localize the independent genetic effect in the IL2/IL21 locus in lupus.
Results
We established and confirmed the genetic association between IL2/IL21 and lupus. Using conditional analysis and trans-ethnic mapping, we localized the genetic effect in this locus to two SNPs in high linkage disequilibrium; rs907715 located within IL21 (OR=1.16 (1.10–1.22), P= 2.17 ×10−8), and rs6835457 located in the 3’-UTR flanking region of IL21 (OR= 1.11 (1.05–1.17), P= 9.35×10−5).
Conclusion
We have established the genetic association between lupus and IL2/IL21 with a genome-wide level of significance. Further, we localized this genetic association within the IL2/IL21 linkage disequilibrium block to IL21. If other autoimmune IL2/IL21 genetic associations are similarly localized, then the IL21 risk alleles would be predicted to operate in a fundamental mechanism that influences the course of a number of autoimmune disease processes.
Introduction
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease characterized by the production of autoantibodies against nuclear self-antigens. While the etiology of lupus is not certain, a number of genetic susceptibility loci have been identified. (1–2) We have previously reported genetic association between common variants within the IL21 gene and lupus. (3) The IL2/IL21 region at 4q27 has been identified as a genetic susceptibility locus in a number of autoimmune disorders.(4–8) While these data suggest that the IL2/IL21 region is a genetic susceptibility locus for human autoimmunity, the localization of this genetic effect at this locus has not been established. Indeed, both IL2 and IL21 are equally attractive biological candidates.
Aberrant regulation of both IL-2 and IL-21 has been implicated as a potential driver of autoimmunity in human and murine lupus. (9–11) IL-2 plays an important role in regulatory T cell homeostasis and survival but is not essential for Treg proliferation or suppression. (12–13) However, reduced survival of regulatory T cells in response to reduced IL-2 expression has been observed, leading to autoimmunity in NOD mice. (14) IL-21 is a cytokine that plays a central role in the antibody mediated immune response. Produced primarily by CD4+ T cells, it acts on NK cells, CD4+ T cells, and B lymphocytes to induce and sustain antibody production and mediate antibody class switching. (15) IL-21 also induces Th17 differentiation through a pathway distinct from IL-6 involving STAT3 signaling and activation of RORγt.(16–17) Th17 cells are mediators of inflammation and have been shown to possess a pathogenic role in autoimmunity.(18) While IL-21 producing CD4+ T cells readily arise under Th17 polarizing conditions, not all CD4+ T cells that produce IL-21 produce IL-17A or IL-17F. (19) IL-6, IL-21 and STAT3 signaling have also been shown to be important in T follicular helper proliferation. (20) An uncontrolled T follicular helper response has been shown to induce systemic autoimmunity through excess germinal center formation and high affinity antibody production. (21–22)
Herein we confirm, replicate, and fine map the genetic association with the IL2/IL21 locus in lupus using two large European-derived and African-American lupus sample sets. We localize the genetic association in this locus, and demonstrate that the main genetic effect in this locus is explained by the genetic association with IL21.
Materials and Methods
Study participants and genotyping
A total of 4,248 lupus cases and 3,818 controls of European descent and 1,569 African-American lupus cases and 1,893 African-American controls were included in this study. 45 haplotype tagging SNPs (tag r2≥0.8) were genotyped in the IL2/IL21 locus at 4q27 to cover the entire LD block in that locus spanning from KIAA1109 to BBS12. Tag SNPs were selected using HapMap CEU data as our European-derived population represents the larger sample set included in this study, and capture all HapMap SNPs in the LD block examined with a mean r2 value of 0.96. In addition, 347 ancestry informative markers (AIMs) were genotyped (23–26). All lupus cases fulfilled the ACR lupus classification criteria (27–28). Genotyping was performed using Illumina Custom Bead system on the iSCAN instrument, as part of a large multi-investigator candidate gene association study for lupus that included a total of 32,216 SNPs in a number of candidate genes. This collaborative genotyping approach helped us to maximize our sample size and reduce genotyping costs.
Data Analysis
Individuals with a genotype success rate of <90% were excluded from the analysis. A total of 326 and 17 European-derived and African-American samples, respectively, were removed due to low genotype success rate. The remaining samples were then evaluated for duplicates or related individuals and one individual from each pair was removed if the proportion of alleles shared identical by descent (IBD) > 0.4. Samples with increased heterozygosity (>5 standard deviation around the mean) were then removed from the analysis. Finally, genetic outliers were removed from further analysis as determined by principal components analysis (PCA) and admixture proportions calculated using ADMIXMAP. A total of 10 principal components were calculated and 2 principal components that explain the majority of variation in samples included in this study were used to identify outliers in the European-derived and African-American sample sets. Outliers were defined by 4 standard deviations from the mean of each of the 2 principal components. Addition outliers identified using ADMIXMAP were also removed. A total of 51 genetic outliers were removed from the European-derived samples (44 outliers identified by PCA and 7 additional outliers identified by ADMIXMAP). In the African-American sample, a total of 30 outliers were removed (25 outliers identified by PCA and 5 additional outliers identified by ADMIXMAP). After filtering based on the aforementioned criteria, a total 3,980 cases and 3,546 control individuals of European descent and 1,414 cases and 1,767 controls of African-American descent were included in subsequent analyses. All study participants singed an informed consent, and all protocols were approved by the institutional review boards of our institutions.
For each sample set analyzed, markers with a genotype success rate (GSR) below 0.90, Hardy-Weinberg equilibrium (HWE) P value below 0.001, or minor allele frequency (MAF) less than 0.01 were excluded from further analysis. Of the 45 markers genotyped, 35 passed the inclusion threshold in European-derived participants and were subsequently analyzed (10 SNPs were excluded: 3 SNPs due to HWE, GSR, and MAF; 3 due to HWE only; 3 due to GSR only; 1 due to MAF only). 35 markers were analyzed in African-Americans (10 SNPs were excluded: 3 due to HWE, GSR, and MAF; 1 due to HWE only; 1 due to GSR only; 5 due to MAF only).
Allele frequencies in patients and controls, odds ratios and corresponding 95 percent confidence intervals, and χ2 with corresponding P values were determined for each SNP using PLINK (29). Hardy-Weinberg equilibrium P values were calculated and LD plots generated using Haploview 4.2 (30).
Pairwise conditional analysis was performed in the European-derived sample set to test for associations independent of the haplotypic background. Two-marker haplotypes were constructed using all associated SNPs. Total haplotypic association was calculated for each two-marker haplotype constructed. Each two-marker haplotype was subsequently conditioned on each of its constituent markers. Markers maintaining significance upon conditioning were said to have an effect independent of the alternate marker in the two-marker haplotype.
Meta-analysis was performed on the single SNP associations obtained for rs6835457 and rs907715 using the European-derived and African-American participants using PLINK. Meta-analysis odds-ratios and corresponding 95 percent confidence intervals, P values, and heterogeneity p values were calculated for both markers.
Imputation analysis was performed using Impute version 2 (31).
Results
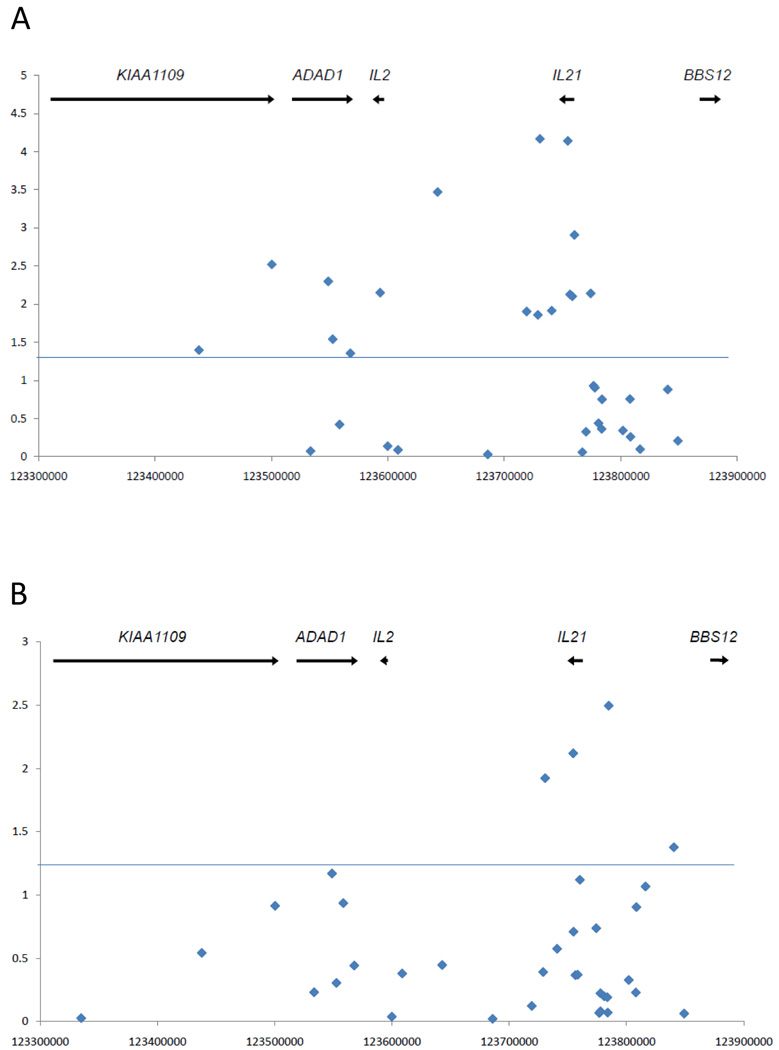
In the European-derived participants, a number of SNPs contained throughout the IL2/IL21 region are significantly associated with lupus susceptibility (P<0.05). (Table 1) Markers in and around IL21 show the most significant association with lupus susceptibility in individuals of European ancestry. (Table 1, Figure 1A) The two markers with the highest association (rs6835457, P= 6.76×10−5; rs907715, P= 7.21×10−5) are located in the IL21 3’-UTR flanking region, and within IL21, respectively. (Figure 1A, 2A) Two SNPs in IL2 show significant association with lupus susceptibility in the European-derived sample set (rs2069779, P=0.0071; rs6814718, P= 0.00034).
Table 1.
Genetic association between IL2/IL21 in European-derived lupus patients and controls.
| Associated | Frequency | 95% CI | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Position | Gene | Function | Allele | Cases | Controls | OR | LL | UL | P Value |
| rs13119723 | 123437763 | KIAA1109 | Intron | A | 0.88 (6633:935) | 0.87 (5840:912) | 1.11 | 1.01 | 1.22 | 0.04 |
| rs1127348 | 123500310 | KIAA1109 | Coding | G | 0.23 (1843:6073) | 0.21 (1507:5581) | 1.12 | 1.04 | 1.21 | 0.003 |
| rs6814233 | 123533582 | ADAD1 | Intron | C | 0.70 (5421:2367) | 0.70 (4826:2122) | 1.01 | 0.94 | 1.08 | 0.85 |
| rs17388568 | 123548812 | ADAD1 | Intron | A | 0.28 (2168:5610) | 0.26 (1795:5157) | 1.11 | 1.03 | 1.19 | 0.005 |
| rs12499753 | 123552627 | ADAD1 | Intron | G | 0.34 (2587:5145) | 0.32 (2205:4737) | 1.08 | 1.01 | 1.16 | 0.029 |
| rs6827839 | 123558465 | ADAD1 | Intron | A | 0.40 (3056:4536) | 0.40 (2678:4096) | 1.03 | 0.96 | 1.10 | 0.38 |
| rs11732095 | 123567795 | ADAD1 | Intron | A | 0.91 (7205:725) | 0.90 (6346:714) | 1.12 | 1.00 | 1.25 | 0.044 |
| rs2069779 | 123593347 | IL2 | Intron | A | 0.08 (586:7002) | 0.07 (444:6324) | 1.19 | 1.05 | 1.36 | 0.0071 |
| rs4833248 | 123599855 | IL2 | Flanking 5' UTR | G | 0.70 (5355:2345) | 0.69 (4754:2108) | 1.01 | 0.94 | 1.09 | 0.73 |
| rs10857092 | 123608669 | IL2 | Flanking 5' UTR | A | 0.07 (519:7433) | 0.06 (456:6632) | 1.02 | 0.89 | 1.16 | 0.82 |
| rs6814718 | 123642766 | IL2 | Flanking 5' UTR | G | 0.73 (5779:2113) | 0.71 (4975:2073) | 1.14 | 1.06 | 1.22 | 0.0003 |
| rs4833834 | 123685801 | IL21 | Flanking 3' UTR | G | 0.10 (816:7070) | 0.10 (725:6309) | 1.00 | 0.90 | 1.12 | 0.94 |
| rs13140464 | 123719195 | IL21 | Flanking 3' UTR | C | 0.86 (6770:1108) | 0.85 (5917:1087) | 1.12 | 1.03 | 1.22 | 0.013 |
| rs6822844 | 123728871 | IL21 | Flanking 3' UTR | C | 0.86 (6829:1121) | 0.85 (5984:1100) | 1.12 | 1.02 | 1.23 | 0.014 |
| rs6835457 | 123730576 | IL21 | Flanking 3' UTR | A | 0.68 (5433:2507) | 0.65 (4629:2453) | 1.15 | 1.07 | 1.23 | 6.8 ×10−5 |
| rs975404 | 123740742 | IL21 | Flanking 3' UTR | A | 0.65 (5169:2745) | 0.63 (4443:2571) | 1.09 | 1.02 | 1.17 | 0.012 |
| rs907715 | 123754503 | IL21 | Intron | G | 0.69 (5374:2462) | 0.66 (4590:2416) | 1.15 | 1.07 | 1.23 | 7.2 ×10−5 |
| rs4833837 | 123756413 | IL21 | Coding | G | 0.32 (2508:5412) | 0.30 (2093:4967) | 1.10 | 1.03 | 1.18 | 0.0074 |
| rs2221903 | 123758362 | IL21 | Intron | G | 0.32 (2474:5382) | 0.30 (2063:4935) | 1.10 | 1.03 | 1.18 | 0.0079 |
| rs13143866 | 123760208 | IL21 | Intron | G | 0.74 (5523:1985) | 0.71 (4733:1921) | 1.13 | 1.05 | 1.22 | 0.0012 |
| rs4295278 | 123766991 | IL21 | Flanking 5' UTR | A | 0.95 (7563:375) | 0.95 (6734:338) | 1.01 | 0.87 | 1.18 | 0.87 |
| rs4833838 | 123770148 | IL21 | Flanking 5' UTR | A | 0.97 (7690:258) | 0.97 (6841:245) | 1.07 | 0.89 | 1.28 | 0.47 |
| rs6840978 | 123774157 | IL21 | Flanking 5' UTR | G | 0.82 (6553:1399) | 0.81 (5714:1366) | 1.12 | 1.03 | 1.22 | 0.0072 |
| rs13137822 | 123776686 | IL21 | Flanking 5' UTR | C | 0.45 (3498:4334) | 0.43 (3043:3971) | 1.05 | 0.99 | 1.12 | 0.12 |
| rs2137497 | 123777704 | IL21 | Flanking 5' UTR | C | 0.58 (4564:3252) | 0.58 (4035:2917) | 1.01 | 0.95 | 1.08 | 0.66 |
| rs2390352 | 123777780 | IL21 | Flanking 5' UTR | A | 0.96 (7530:336) | 0.95 (6691:337) | 1.13 | 0.97 | 1.32 | 0.13 |
| rs7694252 | 123780886 | IL21 | Flanking 5' UTR | A | 0.71 (5543:2277) | 0.70 (4879:2071) | 1.03 | 0.96 | 1.11 | 0.37 |
| rs6419221 | 123783569 | IL21 | Flanking 5' UTR | G | 0.64 (5048:2806) | 0.64 (4503:2571) | 1.03 | 0.96 | 1.10 | 0.43 |
| rs1533236 | 123783908 | IL21 | Flanking 5' UTR | G | 0.51 (4060:3896) | 0.50 (3541:3551) | 1.04 | 0.98 | 1.11 | 0.18 |
| rs10518400 | 123801876 | IL21 | Flanking 5' UTR | A | 0.08 (600:7354) | 0.07 (512:6576) | 1.05 | 0.93 | 1.18 | 0.45 |
| rs9307509 | 123807941 | IL21 | Flanking 5' UTR | T | 0.91 (6673:639) | 0.91 (5947:617) | 1.08 | 0.96 | 1.22 | 0.18 |
| rs6837455 | 123808392 | IL21 | Flanking 5' UTR | C | 0.15 (1189:6755) | 0.15 (1036:6050) | 1.03 | 0.94 | 1.13 | 0.55 |
| rs13147359 | 123816621 | IL21 | Flanking 5' UTR | A | 0.97 (7691:229) | 0.97 (6847:209) | 1.02 | 0.85 | 1.24 | 0.80 |
| rs309414 | 123840362 | BBS12 | Flanking 3' UTR | A | 0.39 (2974:4606) | 0.38 (2619:4271) | 1.05 | 0.98 | 1.13 | 0.13 |
| rs17006053 | 123849111 | BBS12 | Flanking 3' UTR | C | 0.76 (5958:1836) | 0.76 (5275:1657) | 1.02 | 0.94 | 1.10 | 0.62 |
OR, odds ratio; LL, lower limit; UL, upper limit; CI, confidence interval. The numbers between parenthesis represent the number of risk (associated) and protective (non-associated) alleles, respectively.
Figure 1.
Genetic association at the IL2/IL21 locus in the European-derived (A), and the African-American (B) sample sets included in this study. Y-axis values represent −log(10)-p-values for individual markers genotyped, while x-axis values represent chromosomal coordinates for each marker.
Figure 2.
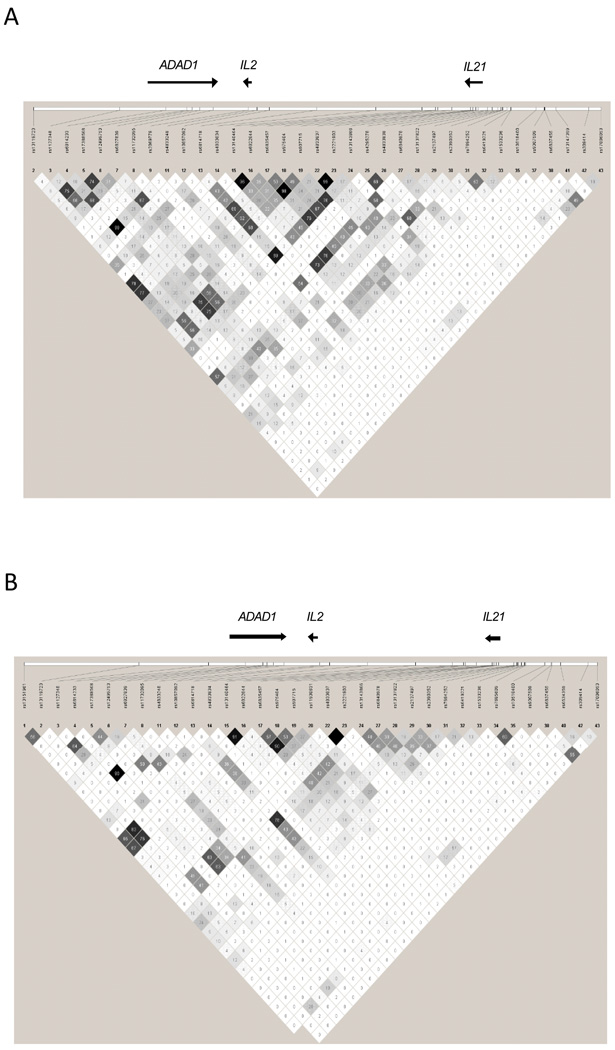
LD plots depicting genetic markers analyzed in the European-derived (A) and the African-American (B) participants included in this study. Pairwsie r2 values are shown.
Pairwise conditional analyses were performed on significantly associated markers in the European-derived sample set. The strongest genetic effect observed in rs6835457 maintained significance when conditioned against all other associated markers. However, rs6835457 located in the 3’-UTR flanking region of IL21 is in almost complete LD with rs907715, a SNP located within IL21 (r2=0.98). This made it impossible to distinguish an independent effect of these SNPs using conditional analysis, and therefore either of the two IL21 SNPs or both can explain the genetic association between lupus and the IL2/IL21 LD block. (Table 2)
Table 2.
Pairwise two-SNP haplotype conditional analysis of rs6835457 with all other associated SNPs in the IL2/IL21 locus in the European-derived lupus patients and controls.
| Single SNP | Haplotype | Conditional Marker P Value | ||
|---|---|---|---|---|
| Marker | P Value | P Value | Haplotype SNP | rs6835457 |
| rs13119723 | 0.04 | 0.00071 | 0.0038 | 0.60 |
| rs1127348 | 0.003 | 0.0001 | 0.0016 | 0.12 |
| rs17388568 | 0.005 | 0.00016 | 0.0024 | 0.18 |
| rs12499753 | 0.029 | 0.00059 | 0.0019 | 0.44 |
| rs11732095 | 0.044 | 0.00036 | 0.00042 | 0.95 |
| rs2069779 | 0.0071 | 0.000044 | 0.00055 | 0.034 |
| rs6814718 | 0.0003 | 0.00090 | 0.19 | 0.63 |
| rs13140464 | 0.013 | 0.00035 | 0.0037 | 0.58 |
| rs6822844 | 0.014 | 0.00035 | 0.0018 | 0.84 |
| rs6835457 | 0.000068 | -- | -- | -- |
| rs975404 | 0.012 | 0.00086 | 0.0052 | 0.64 |
| rs907715 | 0.000072 | 0.00011 | NA | NA |
| rs4833837 | 0.0074 | 0.00024 | 0.0016 | 0.39 |
| rs2221903 | 0.0079 | 0.00023 | 0.0020 | 0.33 |
| rs13143866 | 0.0012 | 0.00064 | 0.10 | 0.85 |
| rs6840978 | 0.0072 | 0.00036 | 0.0042 | 0.89 |
There is, however, a smaller effect in IL2 that is not entirely accounted for by the effect in rs6835457 and rs907715 in lupus patients of European descent. Specifically, rs2069779 has an effect that is independent of the most significant marker rs6835457, as rs2069779 and rs6835457 are not in strong LD (r2=0.02).
Genetic association in the African-American sample set reveals a total of four associated markers (Table 3). Notably, all significant markers are contained in the IL21 region (Figure 1B, 2B), and the two most significantly lupus-associated SNPs in the European-derived participants are also associated with lupus in African-Americans. Furthermore, the only markers found to possess significant association in both ethnicities are rs6835457 and rs907715, located in the 3’UTR flanking regions of IL21 and within IL21, respectively. Similar to the European-derived participants, rs6835457 and rs907715 have a high degree of LD in the African-Americans, suggesting that they represent the same genetic effect (r2= 0.90).
Table 3.
Genetic association between IL2/IL21 in African-American lupus patients and controls.
| Associated | Frequency | 95% CI | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Position | Gene | Function | Allele | Cases | Controls | OR | LL | UL | P Value |
| rs13151961 | 123334952 | KIAA1109 | Intron | A | 0.96 (2475:103) |
0.96 (3188:134) |
1.01 | 0.78 | 1.31 | 0.94 |
| rs13119723 | 123437763 | KIAA1109 | Intron | G | 0.03 (75:2735) |
0.02 (79:3427) |
1.19 | 0.86 | 1.64 | 0.29 |
| rs1127348 | 123500310 | KIAA1109 | Coding | A | 0.97 (2744:82) |
0.96 (3403:127) |
1.25 | 0.62 | 1.66 | 0.12 |
| rs6814233 | 123533582 | ADAD1 | Intron | A | 0.08 (212:2602) |
0.07 (251:3247) |
1.05 | 0.87 | 1.27 | 0.59 |
| rs17388568 | 123548812 | ADAD1 | Intron | G | 0.96 (2700:124) |
0.95 (3334:190) |
1.24 | 0.98 | 1.56 | 0.068 |
| rs12499753 | 123552627 | ADAD1 | Intron | A | 0.90 (2541:283) |
0.90 (3156:372) |
1.06 | 0.90 | 1.25 | 0.50 |
| rs6827839 | 123558465 | ADAD1 | Intron | A | 0.63 (1769:1035) |
0.61 (2117:1345) |
1.09 | 0.98 | 1.20 | 0.12 |
| rs11732095 | 123567795 | ADAD1 | Intron | A | 0.97 (2732:82) |
0.97 (3411:117) |
1.14 | 0.86 | 1.52 | 0.36 |
| rs4833248 | 123599855 | IL2 | Flanking 5' UTR | A | 0.07 (208:2598) |
0.07 (252:3180) |
1.01 | 0.83 | 1.22 | 0.92 |
| rs10857092 | 123608669 | IL2 | Flanking 5' UTR | A | 0.06 (162:2666) |
0.05 (186:3348) |
1.09 | 0.88 | 1.36 | 0.42 |
| rs6814718 | 123642766 | IL2 | Flanking 5' UTR | G | 0.77 (2179:643) |
0.76 (2691:839) |
1.06 | 0.94 | 1.19 | 0.36 |
| rs4833834 | 123685801 | IL21 | Flanking 3' UTR | G | 0.02 (55:2773) |
0.02 (68:3466) |
1.01 | 0.71 | 1.45 | 0.95 |
| rs13140464 | 123719195 | IL21 | Flanking 3' UTR | C | 0.97 (2726:80) |
0.97 (3380:104) |
1.05 | 0.78 | 1.41 | 0.75 |
| rs6822844 | 123728871 | IL21 | Flanking 3' UTR | C | 0.97 (2745:81) |
0.97 (3418:114) |
1.13 | 0.85 | 1.51 | 0.41 |
| rs6835457 | 123730576 | IL21 | Flanking 3' UTR | A | 0.61 (1730:1096) |
0.58 (2051:1479) |
1.14 | 1.03 | 1.26 | 0.012 |
| rs975404 | 123740742 | IL21 | Flanking 3' UTR | A | 0.62 (1743:1081) |
0.60 (2128:1398) |
1.06 | 0.96 | 1.17 | 0.27 |
| rs907715 | 123754503 | IL21 | Intron | G | 0.62 (1762:1060) |
0.59 (2083:1439) |
1.15 | 1.04 | 1.27 | 0.0076 |
| rs11930631 | 123754745 | IL21 | Intron | A | 0.31 (861:1949) |
0.29 (1014:2466) |
1.07 | 0.96 | 1.20 | 0.20 |
| rs4833837 | 123756413 | IL21 | Coding | A | 0.93 (2629:199) |
0.92 (3267:267) |
1.08 | 0.89 | 1.31 | 0.43 |
| rs2221903 | 123758362 | IL21 | Intron | A | 0.93 (2629:199) |
0.92 (3265:267) |
1.08 | 0.89 | 1.31 | 0.43 |
| rs13143866 | 123760208 | IL21 | Intron | G | 0.79 (2211:579) |
0.77 (2665:779) |
1.12 | 0.99 | 1.26 | 0.076 |
| rs6840978 | 123774157 | IL21 | Flanking 5' UTR | G | 0.86 (2430:394) |
0.85 (2999:535) |
1.10 | 0.96 | 1.27 | 0.18 |
| rs13137822 | 123776686 | IL21 | Flanking 5' UTR | C | 0.70 (1963:851) |
0.70 (2437:1067) |
1.01 | 0.91 | 1.12 | 0.86 |
| rs2137497 | 123777704 | IL21 | Flanking 5' UTR | A | 0.71 (2002:812) |
0.71 (2466:1030) |
1.03 | 0.92 | 1.15 | 0.60 |
| rs2390352 | 123777780 | IL21 | Flanking 5' UTR | G | 0.53 (1500:1326) |
0.53 (1860:1662) |
1.01 | 0.92 | 1.12 | 0.83 |
| rs7694252 | 123780886 | IL21 | Flanking 5' UTR | G | 0.22 615:2197) |
0.21 (751:2763) |
1.03 | 0.91 | 1.16 | 0.63 |
| rs6419221 | 123783569 | IL21 | Flanking 5' UTR | G | 0.61 (1727:1093) |
0.61 (2138:1386) |
1.02 | 0.93 | 1.13 | 0.64 |
| rs1533236 | 123783908 | IL21 | Flanking 5' UTR | G | 0.38 (1070:1758) |
0.38 (1329:2205) |
1.01 | 0.91 | 1.12 | 0.85 |
| rs7685609 | 123784752 | IL21 | Flanking 5' UTR | G | 0.98 (2694:68) |
0.96 (3302:130) |
1.56 | 1.16 | 2.10 | 0.0032 |
| rs10518400 | 123801876 | IL21 | Flanking 5' UTR | G | 0.95 (2668:156) |
0.94 (3220:210) |
1.08 | 0.87 | 1.34 | 0.47 |
| rs9307509 | 123807941 | IL21 | Flanking 5' UTR | T | 0.97 (2599:91) |
0.96 (3122:118) |
1.08 | 0.82 | 1.43 | 0.59 |
| rs6837455 | 123808392 | IL21 | Flanking 5' UTR | C | 0.22 (612:2212) |
0.20 (707:2811) |
1.10 | 0.97 | 1.24 | 0.13 |
| rs6534359 | 123816144 | IL21 | Flanking 5' UTR | A | 0.94 (2651:157) |
0.93 (3277:233) |
1.20 | 0.97 | 1.48 | 0.086 |
| rs309414 | 123840362 | BBS12 | Flanking 3' UTR | C | 0.76 (2147:673) |
0.74 (2600:918) |
1.13 | 1.00 | 1.26 | 0.042 |
| rs17006053 | 123849111 | BBS12 | Flanking 3' UTR | C | 0.75 (2099:689) |
0.75 (2600:862) |
1.01 | 0.90 | 1.13 | 0.87 |
OR, odds ratio; LL, lower limit; UL, upper limit, CI, confidence interval. The numbers between parenthesis represent the number of risk (associated) and protective (non-associated) alleles, respectively.
To establish the genetic association detected in IL21 with a genome-wide significance (P<5× 10−8), we attempted to increase the sample size by including control samples from the Wellcome Trust Case-Control Consortium (WTCCC). Since neither rs6835457 nor rs907715 were genotyped in WTCCC, we imputed genotype calls for rs907715 and rs683545 in the 3,004 WTCCC control individuals from a reference panel consisting of phased HapMap3 and 1000 Genomes haplotypes. Genotype calls were made on the basis of a 0.75 imputation threshold. Imputation success rate was >90% for both imputed SNPs. Imputed calls for each marker were combined with the non-imputed genotypes obtained in our European-derived study participants, and the genetic association test was repeated. Meta-analysis of associations obtained in our European-derived participants combined with WTCCC controls and our African-American study participants was performed. The genetic association between lupus and rs907715 within IL21 was validated and with a genome-wide significance (P= 2.17 ×10−8) establishing the genetic association between the IL21 locus with lupus (Table 4).
Table 4.
Meta-analysis of genetic associations in the European-derived sample set combined with WTCCC controls and the African- American sample set for markers in the IL21 gene.
| Associated Allele |
European-derived with WTCCC Controls |
African-American | Meta-analysis | |||||
|---|---|---|---|---|---|---|---|---|
| SNP | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | Pheterogeneity | |
| rs6835457 | A | 1.10 (1.03–1.17) | 2.3×10−3 | 1.14 (1.03 1.26) | 0.012 | 1.11 (1.05–1.17) | 9.35×10−5 | 0.55 |
| rs907715 | G | 1.16 (1.04–1.27) | 8.41×10−7 | 1.15 (1.04 1.27) | 0.0076 | 1.16 (1.10–1.22) | 2.17 ×10−8 | 0.83 |
Discussion
Our data confirmed, replicated and localized the genetic association between the IL2/IL21 haplotype block and susceptibility to lupus. We observed genetic association in both IL2 and IL21 genes in the European-derived participants with the most significant association residing in and around IL21. In the European-derived sample set there is also a smaller but independent association with one marker contained within the IL2 gene. Analysis in the African-American participants revealed far fewer significant SNPs associated with lupus than in the European-derived sample set, likely reflecting smaller haplotype blocks in African-Americans. Interestingly, the two most significantly associated markers in the European-derived sample set (contained in and around IL21) were also associated with lupus in the African-American participants, while polymorphisms in IL2 were not associated with lupus in African-Americans (Table 2). Although rs2069779 located in IL2, did not meet the inclusion criterion for minor allele frequency in African-Americans (rs2069779, MAF= 0.9%), no difference in allele frequency was observed between cases and controls (MAFCase= 0.009, MAFControl= 0.009). While the genetic association with rs2069779 in IL2 appears to be independent of the main genetic association in this locus in IL21 in the European-derived sample set, this effect was not reproducible in the African-American lupus patients. The failure to confirm this IL2 association in African-Americans further supports a stronger role for IL21 polymorphisms in lupus susceptibility at the IL2/IL21 locus.
We and others have demonstrated the genetic association of polymorphisms within the IL2/IL21 locus at 4q27 with lupus and multiple other autoimmune and inflammatory disorders. (3–7, 32–33) Our data show a modest association between lupus and the commonly studied IL2/IL21 inter-genic marker rs6822844 in the European-derived but not the African-American lupus sample set (P= 0.014, 0.41, respectively). These data, together with conditional analysis in the European-derived participants, suggest that the observed association between lupus and rs6822844 is explained by the association with rs6835457 and rs907715, which shows an independent genetic effect in our studies. Neither rs6835457 nor rs907715 were included in commonly used genome-wide association platforms and therefore were not evaluated in genome-wide associated studies of other autoimmune or inflammatory diseases.
The association of polymorphisms at 4q27 has been demonstrated in type I diabetes in human studies.(4, 32) Significant evidence for involvement of IL2 and IL21 in type 1 diabetes also comes from the NOD mouse in which the most highly associated non-MHC locus is the Idd3 region of the murine genome which contains the IL2 and IL21 genes. In one study, the overexpression of IL-21 was observed to correlate with the number of Idd3 alleles, and this change in expression was shown to occur in response to polymorphisms which establish an Sp1 binding site upstream of IL21. (34) This study observed no difference in the expression levels of IL-2. In contrast to these findings, a previous study reported the underexpression of IL-2 at the NOD Idd3 locus, leading to the loss of stability in peripheral Treg cell cohorts, and these changes were unaccompanied by changes in IL-21 expression. (14) This reduction in Treg cell numbers was in turn shown to lead to an increase in presentation of beta cell antigens by dendritic cells.
While these studies and others present compelling evidence for the involvement of both IL2 and IL21 in type 1 diabetes, the associations observed in other autoimmune diseases are illustrative as well. Significant epistasis has been reported between rs6822844 and polymorphisms in IL23R in ulcerative colitis.(35) The association of these distal variants suggests a role for polymorphisms at 4q27 in the establishment of a Th17 defect.(36) Furthermore, these data suggest that IL-21, a driver of Th17 differentiation (37), likely accounts for the genetic effect observed at 4q27 in ulcerative colitis.
We have previously reported the association of polymorphisms in IL21R with lupus susceptibility (rs3093301, Pmeta=0.0001, OR=1.16 [95% confidence interval 1.08–1.25]) (38). The association of polymorphisms in the IL21R gene located at 16p11 further implicates the IL-21/IL-21R pathway signaling in lupus risk. IL-21/IL-21R signaling plays a pathogenic role in multiple models of murine lupus. Blockade of IL-21R signaling with IL-21R.Fc attenuates the severity of disease in the MRL/lpr lupus mouse model. (39) The deletion of IL-21R in BXSB-Yaa mice ameliorates antibody-mediated disease manifestations, while IL-21R competent BXSB-Yaa mice produce high levels of IgG1, IgG2b, and IgG3.(10) These data further support a role for an IL-21/IL-21R signaling defect in lupus pathogenesis.
In conclusion, using two large ethnically-diverse lupus sample sets, conditional analysis, and trans-ethnic genotyping, we fine-mapped and localized the genetic association with lupus in the IL2/IL21 LD block to IL21. These data might be relevant to a number of other autoimmune and inflammatory diseases with a reported genetic association in the same region.
Acknowledgements
This work was made possible by NIH Grant Number R03AI076729 from the National Institute of Allergy and Infectious Diseases, NIH Grants Number P20RR020143 and P30AR053483, funding from the Arthritis National Research Foundation and the University of Oklahoma College of Medicine (AHS); NIH Grants Number AR42460, AI024717, AI31584, AR62277, AR048940, AR0490084, Kirkland Scholar award, Alliance for Lupus Research, and US Department of Veterans Affairs (JBH); NIH grant number P01AR049084 (RPK, EEB, SKN, RRG, JBH); NIH grant number AR043274 (KLM); NIH grant number AI063274 (PMG); NIH grant number K24AR002138, P602AR30692, and UL1RR025741 (RRG); NIH grant number R01AR043727 (MP). Funding was also provided by the European Science Foundation to the BIOLUPUS network (07-RNP-083), the Swedish Research Council, the Swedish Association against Rheumatism, the Instituto de Salud Carlos III and Consejería de Salud, Andalucía (MEAR). We are thankful to Dr. Peter Gregersen for providing DNA control samples for our study.
The BIOLUPUS network is composed of: Johan Frostegård, MD, PhD (Huddinge, Sweden), Lennart Truedsson, MD, PhD (Lund, Sweden), Enrique de Ramón, MD PhD (Málaga, Spain), José M. Sabio, MD, PhD (Granada, Spain), María F. González-Escribano, PhD (Sevilla, Spain), Norberto Ortego-Centeno (Granada, Spain), José Luis CAllejas MD (Granada, Spain), Julio Sánchez-Román, MD (Sevilla, Spain), Sandra D’Alfonso, PhD (Novara, Italy), Sergio Migliarese MD (Napoli, Italy), Gian-Domenico Sebastiani MD (Rome, Italy), Mauro Galeazzi MD (Siena, Italy), Torsten Witte, MD, PhD (Hannover, Germany), Bernard R. Lauwerys, MD, PhD (Louvain, Belgium), Emoke Endreffy, PhD (Szeged, Hungary), László Kovács, MD, PhD (Szeged, Hungary), Carlos Vasconcelos, MD, PhD (Porto, Portugal), Berta Martins da Silva, PhD (Porto, Portugal).
Footnotes
None of the authors have any financial conflict of interest with the material presented in this manuscript
References
- 1.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10(5):373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham RR, Hom G, Ortmann W, Behrens TW. Review of recent genome-wide association scans in lupus. J Intern Med. 2009;265(6):680–688. doi: 10.1111/j.1365-2796.2009.02096.x. [DOI] [PubMed] [Google Scholar]
- 3.Sawalha AH, Kaufman KM, Kelly JA, Adler AJ, Aberle T, Kilpatrick J, et al. Genetic association of interleukin-21 polymorphisms with systemic lupus erythematosus. Ann Rheum Dis. 2008;67(4):458–461. doi: 10.1136/ard.2007.075424. [DOI] [PubMed] [Google Scholar]
- 4.Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81(6):1284–1288. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Festen EA, Goyette P, Scott R, Annese V, Zhernakova A, Lian J, et al. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut. 2009;58(6):799–804. doi: 10.1136/gut.2008.166918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39(7):827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Helms C, Liao W, Zaba LC, Duan S, Gardner J, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4(3) doi: 10.1371/journal.pgen.1000041. e1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maiti AK, Kim-Howard X, Viswanathan P, Guillen L, Rojas-Villarraga A, Deshmukh H, et al. Confirmation of an association between rs6822844 at the Il2–Il21 region and multiple autoimmune diseases: evidence of a general susceptibility locus. Arthritis Rheum. 2010;62(2):323–329. doi: 10.1002/art.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dauphinee MJ, Kipper SB, Wofsy D, Talal N. Interleukin 2 deficiency is a common feature of autoimmune mice. J Immunol. 1981;127(6):2483–2487. [PubMed] [Google Scholar]
- 10.Bubier JA, Sproule TJ, Foreman O, Spolski R, Shaffer DJ, Morse HC, 3rd, et al. A critical role for IL-21 receptor signaling in the pathogenesis of systemic lupus erythematosus in BXSB-Yaa mice. Proc Natl Acad Sci U S A. 2009;106(5):1518–1523. doi: 10.1073/pnas.0807309106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ettinger R, Kuchen S, Lipsky PE. Interleukin 21 as a target of intervention in autoimmune disease. Ann Rheum Dis. 2008;67 Suppl 3:iii83–iii86. doi: 10.1136/ard.2008.098400. [DOI] [PubMed] [Google Scholar]
- 12.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol. 2005;6(11):1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 13.D'Cruz LM, Klein L. Development and function of agonist-induced CD25+Foxp3+ regulatory T cells in the absence of interleukin 2 signaling. Nat Immunol. 2005;6(11):1152–1159. doi: 10.1038/ni1264. [DOI] [PubMed] [Google Scholar]
- 14.Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, et al. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet. 2007;39(3):329–337. doi: 10.1038/ng1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parrish-Novak J, Dillon SR, Nelson A, Hammond A, Sprecher C, Gross JA, et al. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature. 2000;408(6808):57–63. doi: 10.1038/35040504. [DOI] [PubMed] [Google Scholar]
- 16.Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448(7152):484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448(7152):480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 18.Pernis AB. Th17 cells in rheumatoid arthritis and systemic lupus erythematosus. J Intern Med. 2009;265(6):644–652. doi: 10.1111/j.1365-2796.2009.02099.x. [DOI] [PubMed] [Google Scholar]
- 19.Suto A, Kashiwakuma D, Kagami S, Hirose K, Watanabe N, Yokote K, et al. Development and characterization of IL-21-producing CD4+ T cells. J Exp Med. 2008;205(6):1369–1379. doi: 10.1084/jem.20072057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29(1):138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435(7041):452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 22.Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, et al. Follicular helper T cells are required for systemic autoimmunity. J Exp Med. 2009;206(3):561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang N, Li H, Criswell LA, Gregersen PK, Alarcon-Riquelme ME, Kittles R, et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005;118(3–4):382–392. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]
- 24.Tian C, Hinds DA, Shigeta R, Adler SG, Lee A, Pahl MV, et al. A genomewide single-nucleotide-polymorphism panel for Mexican American admixture mapping. Am J Hum Genet. 2007;80(6):1014–1023. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30(1):69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez E, Webb RD, Rasmussen A, Kelly JA, Riba L, Kaufman KM, et al. Genetically determined amerindian ancestry correlates with increased frequency of risk alleles for systemic lupus erythematosus. Arthritis Rheum. 2010;62(12):3722–3729. doi: 10.1002/art.27753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 28.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39(7):857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marquez A, Orozco G, Martinez A, Palomino-Morales R, Fernandez-Arquero M, Mendoza JL, et al. Novel association of the interleukin 2-interleukin 21 region with inflammatory bowel disease. Am J Gastroenterol. 2009;104(8):1968–1975. doi: 10.1038/ajg.2009.224. [DOI] [PubMed] [Google Scholar]
- 34.McGuire HM, Vogelzang A, Hill N, Flodstrom-Tullberg M, Sprent J, King C. Loss of parity between IL-2 and IL-21 in the NOD Idd3 locus. Proc Natl Acad Sci U S A. 2009;106(46):19438–19443. doi: 10.1073/pnas.0903561106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glas J, Stallhofer J, Ripke S, Wetzke M, Pfennig S, Klein W, et al. Novel genetic risk markers for ulcerative colitis in the IL2/IL21 region are in epistasis with IL23R and suggest a common genetic background for ulcerative colitis and celiac disease. Am J Gastroenterol. 2009;104(7):1737–1744. doi: 10.1038/ajg.2009.163. [DOI] [PubMed] [Google Scholar]
- 36.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8(9):967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 37.Yang L, Anderson DE, Baecher-Allan C, Hastings WD, Bettelli E, Oukka M, et al. IL-21 and TGF-beta are required for differentiation of human T(H)17 cells. Nature. 2008;454(7202):350–352. doi: 10.1038/nature07021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb R, Merrill JT, Kelly JA, Sestak A, Kaufman KM, Langefeld CD, et al. A polymorphism within IL21R confers risk for systemic lupus erythematosus. Arthritis Rheum. 2009;60(8):2402–2407. doi: 10.1002/art.24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herber D, Brown TP, Liang S, Young DA, Collins M, Dunussi-Joannopoulos K. IL-21 has a pathogenic role in a lupus-prone mouse model and its blockade with IL-21R.Fc reduces disease progression. J Immunol. 2007;178(6):3822–3830. doi: 10.4049/jimmunol.178.6.3822. [DOI] [PubMed] [Google Scholar]