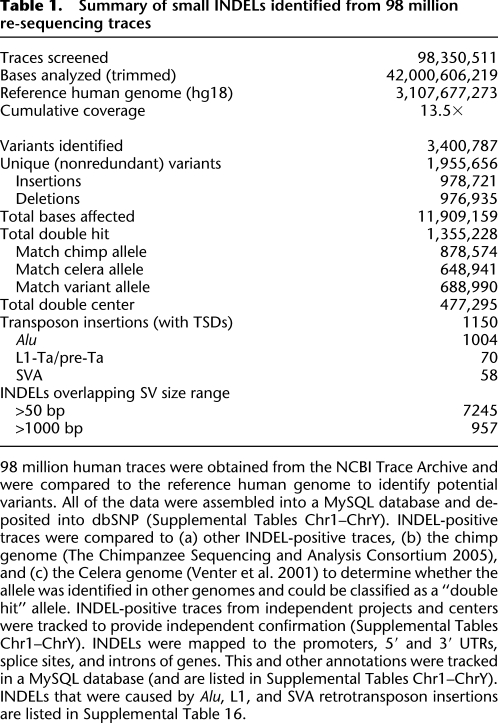
Table 1.
Summary of small INDELs identified from 98 million re-sequencing traces
98 million human traces were obtained from the NCBI Trace Archive and were compared to the reference human genome to identify potential variants. All of the data were assembled into a MySQL database and deposited into dbSNP (Supplemental Tables Chr1–ChrY). INDEL-positive traces were compared to (a) other INDEL-positive traces, (b) the chimp genome (The Chimpanzee Sequencing and Analysis Consortium 2005), and (c) the Celera genome (Venter et al. 2001) to determine whether the allele was identified in other genomes and could be classified as a “double hit” allele. INDEL-positive traces from independent projects and centers were tracked to provide independent confirmation (Supplemental Tables Chr1–ChrY). INDELs were mapped to the promoters, 5′ and 3′ UTRs, splice sites, and introns of genes. This and other annotations were tracked in a MySQL database (and are listed in Supplemental Tables Chr1–ChrY). INDELs that were caused by Alu, L1, and SVA retrotransposon insertions are listed in Supplemental Table 16.