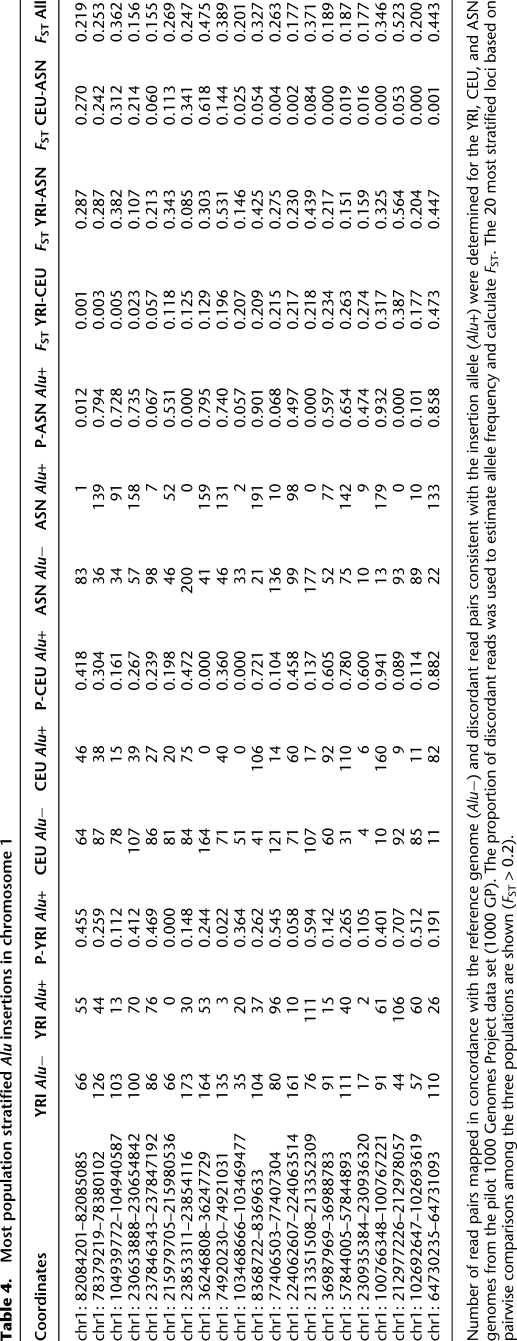
Table 4.
Most population stratified Alu insertions in chromosome 1
Number of read pairs mapped in concordance with the reference genome (Alu−) and discordant read pairs consistent with the insertion allele (Alu+) were determined for the YRI, CEU, and ASN genomes from the pilot 1000 Genomes Project data set (1000 GP). The proportion of discordant reads was used to estimate allele frequency and calculate FST. The 20 most stratified loci based on pairwise comparisons among the three populations are shown (FST > 0.2).