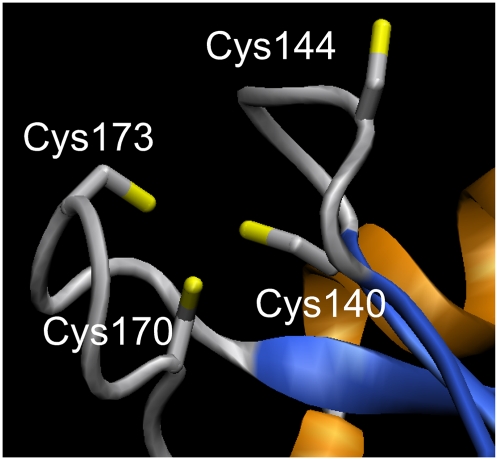
Figure 2.
Putative Zn+2-binding site in target 12087b formed by Cys140 (metal = 0.84; free = 0.02; disulf = 0.14), Cys144 (metal = 0.55; free = 0.15; disulf = 0.3), Cys170 (metal = 0.98; free = 0; disulf = 0.02) and Cys173 (metal = 0.93; free = 0.01; disulf = 0.06). Model obtained with SWISS-MODEL in alignment mode (Kiefer et al. 2009). As a template, we used the Zn+2-binding crystal structure of the Sir2 protein from Archaeoglobus fulgidus (PDB ID: 1ICI) (Min et al. 2001), which shares 22% sequence identity with target 12087b (alignment length is 272, using three iterations of PSI-BLAST). VMD (Humphrey et al. 1996) was used for molecular graphics.