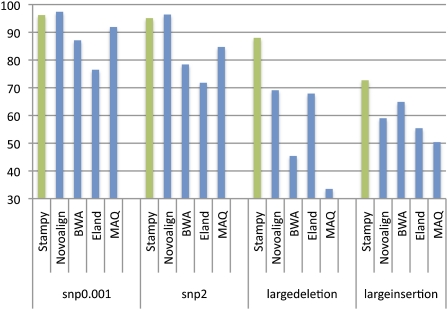
Figure 1.
Recall rates for four sets of 2 million simulated 72-bp paired-end reads, mapped back to the human reference by five read mapping algorithms. Reads included errors following an empirical distribution, as well as additional simulated polymorphisms: 0.1% single nucleotide variants (snp0.001), two single nucleotide variants per read (snp2), and a single large deletion or insertion per read pair (largedeletion and largeinsertion). For details of the simulation procedure, see Supplemental material.