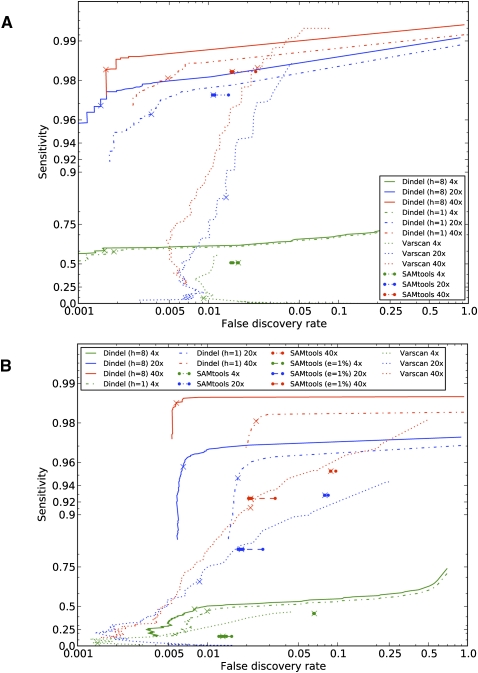
Figure 4.
Accuracy of detection of indel sites of Dindel, SAMtools, and VarScan on simulated data. (A) Sensitivity and false discovery rates for reads simulated with a constant sequencing error indel rate of 0.005% per-base at coverages of 4×, 20×, and 40×. Dindel was run with one candidate haplotype (“h = 1”) and eight candidate haplotypes (“h = 8”). The crosses indicate performance at the 99% confidence level (quality score of 20) of a non-reference indel variant being present. True-positives here are defined as indel calls that result in the same alternative haplotype sequence as that of the simulated indel. (B) Performance on data simulated with indels that were called from high-coverage real data of HapMap individual NA19240 and a realistic sequencing error indel model. Under this model, reads were simulated with increased sequencing error indel rates in long homopolymers, with rates estimated from the low-coverage data set of the 1000 Genomes pilot project. SAMtools was run with a constant sequencing error indel rate of 0.01% and of 1% (“e = 1%”).