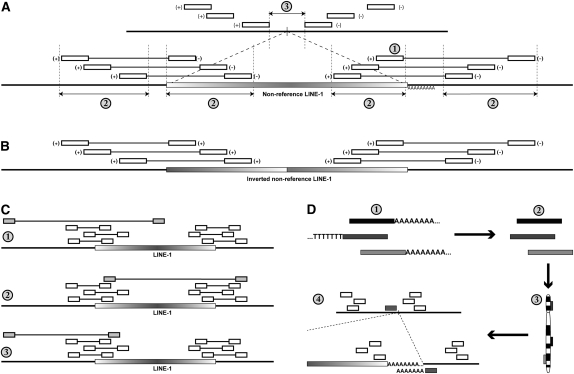
Figure 3.
Bioinformatic procedures for identifying nonreference L1 insertions from whole-genome resequencing data. (Open boxes) Mapped reads indicating the presence of a nonreference L1; (gradient boxes) nonreference L1 insertions; (thicker horizontal lines) genomic sequence. (A) Identification of a nonreference L1 insertion from short-insert paired-end sequence reads. Short-insert paired-end reads where one end matches the reference genome and the other matches an L1 reference are clustered based on mapping location to the human genome reference assembly (top). The criteria for detection as discussed in Methods are labeled with numbers: (1) The 3′ end of the L1 insertion must be represented. (2) Reads must form tight clusters based on the locations of reads mapping to both the reference genome and the reference L1. (3) The minimum distance between the locations of genomic reads must be <100 bp, this interval contains the L1 insertion site (vertical bar). The orientation of the reads is annotated next to the open boxes representing the mapped read positions. (B) L1 insertions may be inverted on the 5′ end (Ostertag and Kazazian 2001), resulting in reads aligning to the reference L1 in the same orientation at the 5′ and 3′ ends of the L1 element. (C) Examples of outlier reads that are filtered as described in Methods. (1) The shaded paired read is an outlier because the locations of the reads corresponding to the L1 and the reference genome do not satisfy criteria 2 in panel A. (2) The shaded paired read is an outlier in terms of the reference L1 location. (3) The location of the shaded paired read is an outlier in terms of the reference genome relative to other reads in the cluster. (D) Identifying reads corresponding to the 3′ junction between the L1 poly-A tail and the reference genome sequence. Reads with 5′ or 3′ poly-T or poly-A stretches of at least six bases (1) are trimmed (2) and aligned to the reference genome assembly (3). Trimmed reads aligning to locations within the predicted L1 insertion (A, 3) site are identified (4).