Figure 7.



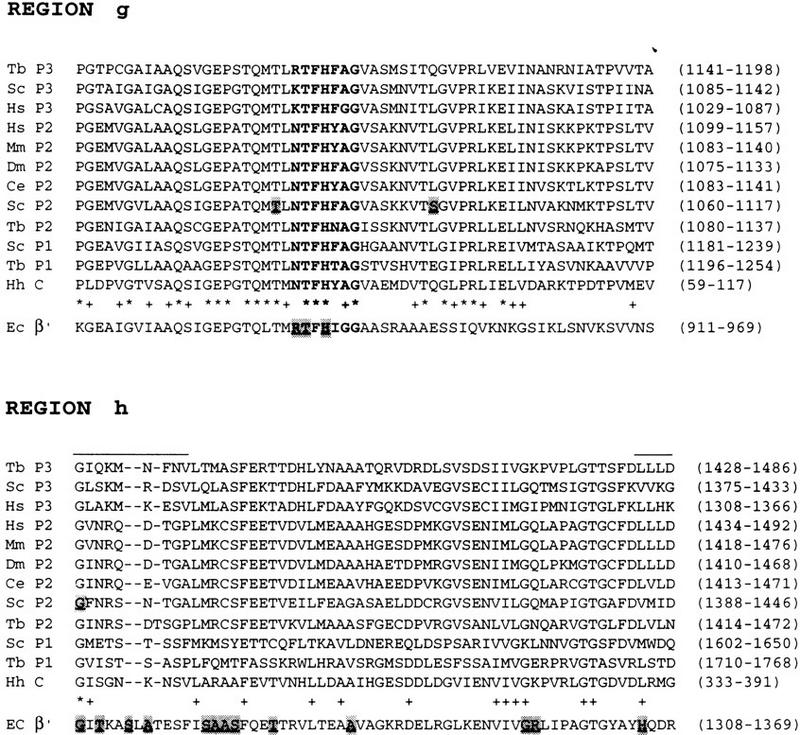
Sequence alignment of the conserved regions a–h of various largest subunits (see p. 1014–1015). The sequence alignments were performed with the CLUSTAL W (v. 1.6) program (Thompson et al. 1994) with the default parameter settings, except for the alignments of the E. coli β′ subunit in the c, f, g, and h regions, which were according to Weilbaecher et al. (1994). Asterisks (*) indicate identities in the 11 eukaryotic sequences; pluses (+) indicate at least 11 out of 12 similarities or identities. The following amino acids were considered similar: V, I, L, and M; D and E; N and Q; R, K, and H; S and T; W, F, and Y. The numbers in parentheses correspond to the first and last amino acids of the sequence shown. The abbreviations for the various organisms are as in Fig. 2. The overlined sequences in regions c, f, and h are not part of the conserved regions and were not included in the percent calculations in Fig. 6. The shaded and underlined residues in the E. coli (Ec) β′ sequences in regions c, f, g, and h correspond to the locations of mutations that affect transcription termination (Weilbaecher et al. 1994). In region a, the conserved amino acids in the zinc DNA binding consensus motif (bracketed) are indicated in boldface type. In region c, sequences from T7 DNA polymerase and E. coli DNA polymerase I have been aligned as described in Allison et al. (1985). Identities and similarities between either of the DNA polymerase sequences and the human (Hs) P3 sequence are indicated by colons. In the S. cerevisiae (Sc) P2 sequence, A402 was mutated in a sua8 suppressor (A402R) (Berroteran et al. 1994). In region d, an invariant motif in all multimeric RNA polymerases is indicated above the alignment, with the D residues involved in Mg+ binding (Zaychikov et al. 1996) in boldface type. The shaded and underlined residues in the Sc P3 sequence correspond to the positions of lethal substitutions; the overlined residues correspond to the positions of single or double (indicated by a bracket) conditional mutants (Dieci et al. 1995). N445 was mutated in a sua8 suppressor (N445S) (Berroteran et al. 1994). In the f region, a putative RNA polymerase III-specific region (Smith et al. 1989a) located upstream of the f region is indicated in boldface type. The isolated residue in bold indicates a position that is occupied by N in all of the α-amanitin-sensitive enzymes, but not in the α-amanitin-resistant enzymes. Residues shaded and underlined in the Sc P3 sequence correspond to the positions of lethal or conditional (indicated in lowercase letters) single, double, or triple alanine scan mutations (Thuillier et al. 1996). The double conditional mutant D829A/R830A in the Sc P3 sequence affects transcription elongation but not transcription complex assembly or formation of the first phosphodiester bond (Thuillier et al. 1996). Residues shaded and underlined in the Mm P2 [L745F, R749P, I779F (Bartolomei and Corden 1995), and N792D (Bartolomei and Corden 1987), mutations], D. melanograster (Dm) P2 [C4 mutation, R741H (Chen et al. 1993)], and C. elegans (Ce) P2 [C777Y/G785E double mutation, cited in Chen et al. (1993)] correspond to the positions of mutations that confer resistance to α-amanitin. In region g, a highly conserved stretch of seven amino acids that is part of the β′ region cross-linked to the 3′ end of nascent RNA (Borukhov et al. 1991) is indicated in boldface type. In the Sc P2 sequence, the shaded and underlined T1080 and S1096 residues indicate the positions of mutations (T1080I and S1096F) that affect start site location (Hekmatpanah and Young 1991). In region h, upstream of the conserved region, the shaded and underlined G residues in the Sc P2 sequence corresponds to the location of a sua8 suppressor (G1388V) (Berroteran et al. 1994).
