Figure 1.
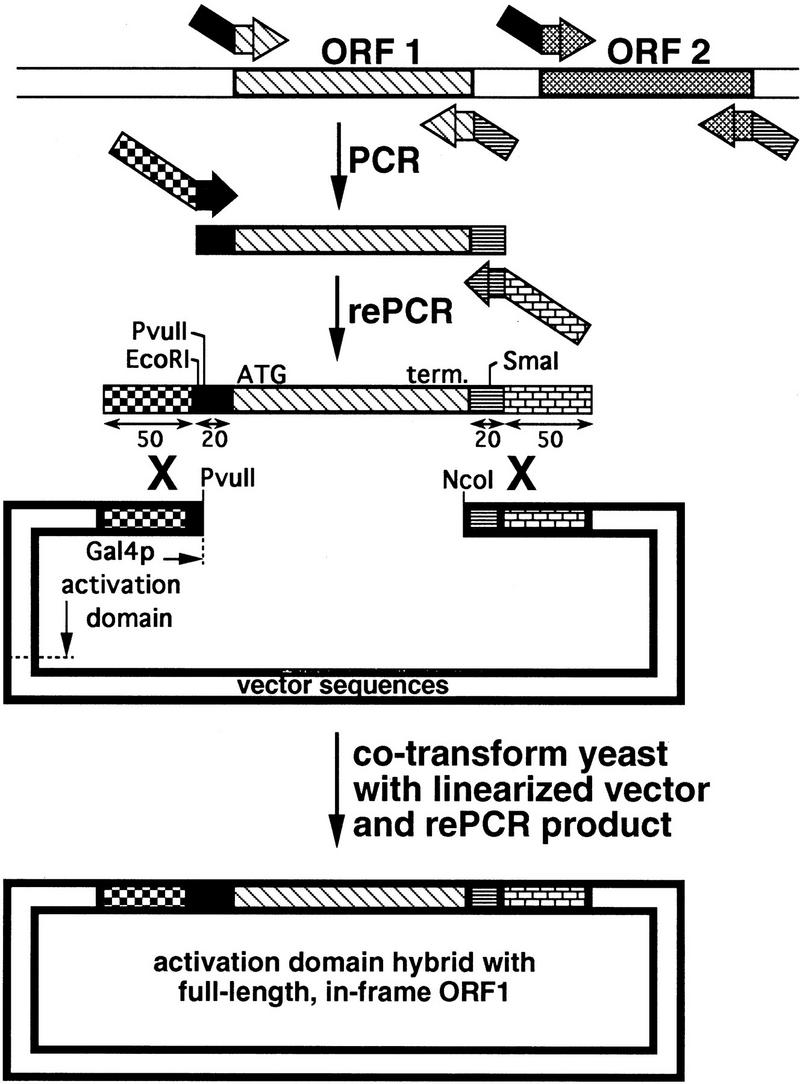
Strategy for cloning each of the yeast ORFs. A segment of the yeast genome containing illustrative ORFs 1 and 2, both of which would be transcribed left to right, is shown at the top. Each ORF is flanked by two bent arrows representing the unique pairs of primers, with the solid black fill (rightward-pointing arrows) indicating the common 5′ termini of the forward primers and the horizontal-lined fill (leftward pointing arrows) indicating the common 5′ termini of the back primers. The sequence of each forward primer is 5′-GGAATTCCAGCTGACCACCATGN17–29-3′ corresponding to 19 bases of non-yeast sequence, the initiator ATG of the ORF, and 17–29 subsequent bases of the ORF. The sequence of each back primer is 5′-GATCCCCGGGAATTGCCATGENDN17–29-3′ corresponding to 20 bases of non-yeast sequence, followed by the reverse complement of a stop codon (noted above as “END”), followed by the reverse complement of 17–29 bases found at the end of the ORF. The product of the PCR of ORF1, shown as a box with diagonal-lined fill flanked by the 19 and 20 bp of non-yeast sequences, is template for the rePCR. The 70-base sequences of the rePCR primers, shown as bent arrows with checkered and “brick” 5′ termini, and 3′ termini matching the sequences in the first set of PCR primers, are provided in Methods. The product of the rePCR is ORF1 flanked by 70-base elements that are identical to those in the two-hybrid vector pOAD. The positions of the translation initiation codon (ATG) and termination codon (term.) of the ORF1 rePCR product are shown. Digestion of the vector with NcoI and PvuII and cotransformation with the rePCR product results in two recombination events that precisely insert the ORF in-frame with the activation domain of Gal4p.
