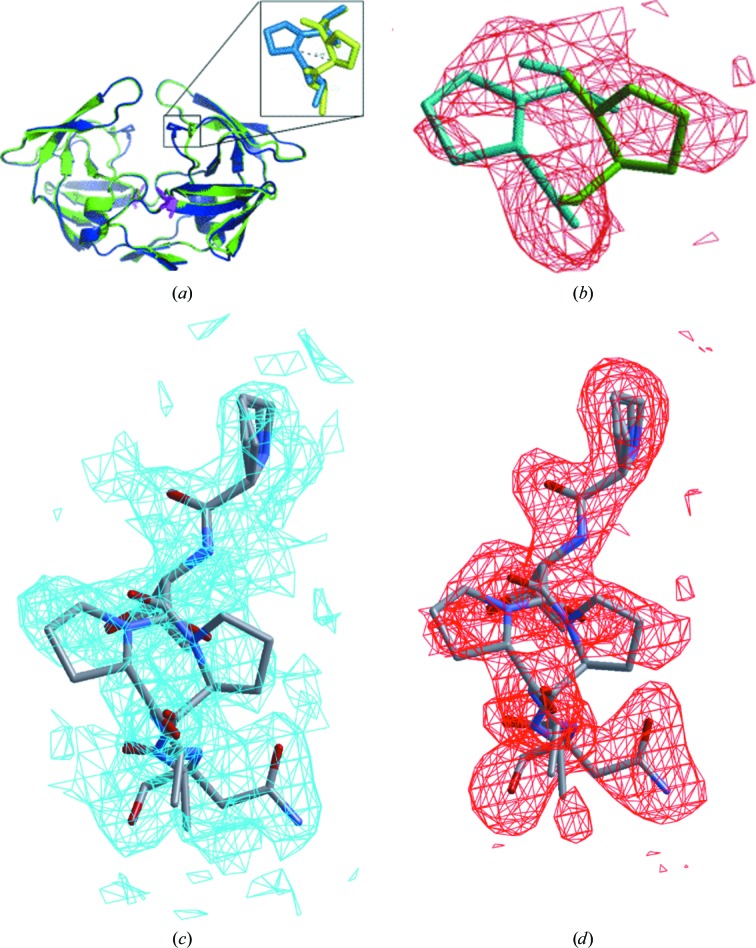
Figure 3.
The proline switch in the MDR769 I10V mutant. (a) MDR769 I10V HIV-1 protease dimer with alternate conformations of Pro81 (shown in the inset). Mutation I10V is highlighted as a stick model in magenta. (b) An |F o| − |F c| OMIT map (contoured at 1.5σ) is shown in red for the alternate conformations of Pro81 in the MDR769 I10V HIV-1 protease crystal structure. Stick models for the alternate conformations of Pro81 with a 3 Å shift in the Cα atoms are shown in the density. The conformation pointing towards the active-site cavity is shown in blue and that pointing away from the active-site cavity is shown in green. (c) 2|F o| − |F c| map (contoured at 1.0σ) for the 80s loop (Pro79–Asn83) including the alternate conformations of Pro81. (d) |F o| − |F c| map (contoured at 1.5σ) for the 80s loop showing the proline switch.