Table 1.
Kinetic parameters of apoMb transient unfolding and refolding measured by R2 relaxation dispersion
| Probes | pH | kN-I1 , s-1 | kI1-N , s-1 | kI1-MG, s-1 | kMG-I1, s-1 | PN | PI1 | PMG |
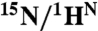 * *
|
5.50 | 38 ± 1 | 495 ± 7 | — | — | 0.93 ± 0.02 | 0.07 ± 0.00 | — |
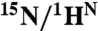 † †
|
4.95 | 31.7 ± 0.5 | 240 ± 12 | 53 ± 1 | 26 ± 4 | 0.71 ± 0.03 | 0.09 ± 0.00 | 0.20 ± 0.03 |
| 3CO/15N‡ | 4.75 | 78 ± 18 | 650 ± 140 | 113 ± 16 | 17 ± 1 | 0.52 ± 0.01 | 0.06 ± 0.01 | 0.42 ± 0.01 |
*Parameters from a two-state fit to amide datasets 1H-SQ and 15N-SQ using the cluster of residues in contact with the F helix and the first helical turn of G (listed in the caption of Fig. S2). Error values were obtained using Monte Carlo sampling.
