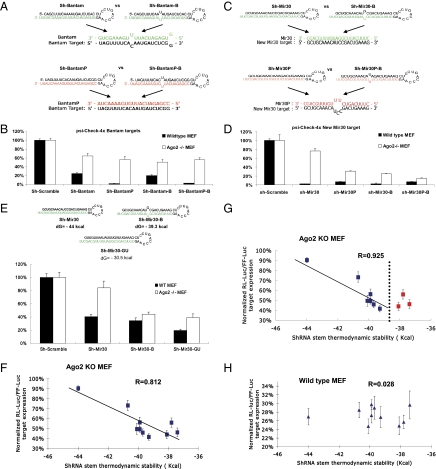
Fig. 4.
RISC function of noncleaving Agos is determined by thermodynamic stability rather than by stem structure of shRNAs. (A) Design of various shRNAs and their associations with targets based on Drosophila miRNA bantam sequence. (B) Wild-type or Ago2 KO MEF cells were cotransfected with the psiCHECK reporter with four tandem bantam target sites in the 3′ UTR and a plasmid expressing various shRNAs. Dual-luciferase assays were performed at 24 h posttransfection, and the results were plotted as described previously. Error bars represent the SD from three independent experiments, each performed in triplicate. (C) Schematic illustration of various shRNAs and interactions between target and guide-strand sequences. Different from Mir30 targets presented in Fig. 2, sh-Mir30 and sh-Mir30-B generate the same guide strand (green), which was a perfect match to the new-mir-30 targets, whereas sh-Mir30P and sh-Mir30P-B produced an identical guide strand (red), which was a mismatch to the new-mir-30 target. (D) psiCHECK vector with four tandem new-mir30 target sites in the 3′ UTR were cotransfected with various shRNA in either wild-type or Ago2 KO MEF cells. Dual-luciferase assays were performed at 24 h posttransfection, and results are plotted as described previously. Error bars represent the SD from three independent experiments, each performed in triplicate transfections. (E) psiCHECK-4xMir30 reporter constructs were cotransfected with the newly designed sh-Mir30-GU, which shares the same guide strand as sh-Mir30 and sh-Mir30-B (green), in either WT or Ago2 KO MEF cells. Results were plotted as described above. Error bars represent the SD from three independent experiments, each performed in triplicate. (F) psiCHECK-4xMir40 reporter constructs were cotransfected with sh-Mir30-3 to sh-Mir30-19 (Fig. S4) individually in Ago2 KO MEF cells. The thermodynamic stability of each shRNA was predicted by mfold (37, 38). The whole sequence, including the loop, was taken into the calculation as an ssRNA. The normalized luciferase reporter level (sh-scramble–treated sample was set as 100%) vs. the shRNA stem thermodynamic stability was plotted. A strong correlation between shRNA-mediated repression efficiency and stem thermodynamic stability was observed (R = 0.812). The more stable the stem, the less repression observed for a particular shRNA via the noncleaving Agos in Ago2 KO MEF cells. More detailed information can be found in Fig. S5 and Table S1. (G) A higher correlation (R = 0.925) was observed when shRNAs with a thermostability lower than a potential threshold (dotted line) were excluded (red squares). (H) The same experiments were performed in wild-type MEF cells. No correlation (R = 0.028) was observed. The results were plotted as described above. Each data point in F and H was the average of 12 trials (four independent experiments, each performed in triplicate) ± SD. The folding energies of shRNAs were also calculated without the stem loop sequences (only mature small RNA duplexes sequences after Dicer processing); the R values were 0.887 and 0.117, respectively.