Abstract
Mutations in different regions of adiponectin gene have been reported to be associated with obesity, atherosclerosis and type 2 diabetes mellitus. The present study was aimed to investigate the association among SNP 45 T > G of adiponectin gene and type 2 diabetes in South Indian population. 75 clinically diagnosed case of type 2 diabetes were studied and compared with 75 apparently healthy controls. The genotype frequency of SNP45 T > G in exon 2 of adiponectin gene was determined by PCR based restriction enzyme analysis using the restriction enzyme SmaI. (recognition site: CCC↓GGG). Three kind of genotypes: wild type TT (470 bp), heterozygous type TG (470 bp, 336 bp, 134 bp) and homozygote mutant type GG (336 bp, 134 bp) were studied. A positive association has been found between SNP45 T > G and type 2 diabetes in the study population (P = 0.010, OR = 3.797, 95% CI = 1.312–10.983). Therefore, SNP45T > G in adiponectin gene may be one of the risk factors for type 2 diabetes.
Keywords: Adiponectin gene, Type 2 diabetes, Polymorphism
Introduction
Diabetes mellitus is a multifactorial, polygenic metabolic disorder which can affect nearly every organ system in the body. According to the Diabetes Atlas 2006 published by the International Diabetes Federation, the number of people with diabetes in India currently around 40.9 million is expected to rise to 69.9 million by 2025 unless urgent preventive steps are taken [1]. In one study [The DESIR (Data from an Epidemiological Study on the Insulin Resistance syndrome) prospective study], 19 common polymorphisms of 14 known candidate genes were analyzed for their contribution to prevalence and incidence of glucose intolerance in middle-aged Caucasian subjects [2]. ADIPOQ is one such gene. The gene is located on chromosome 3q27, a region already known as susceptible to type 2 diabetes and obesity [3]. It spans 17 kb and consists of 3 exons and 2 introns [4]. The gene product is adiponectin which is a protein exclusively secreted from the adipose tissue, mainly the white adipose tissue. Adiponectin has a molecular weight of 30 kDa and composed of 244 amino acids. The protein possesses a short N-terminal variable region followed by collagen repeats and finally a large C-terminal globular domain [5]. It modulates a number of metabolic processes including glucose regulation and fatty acid metabolism by exerting antidiabetic, anti-inflammatory and antiatherogenic effects [6, 7]. Many single nucleotide polymorphisms (SNPs) have been detected in ADIPOQ gene, which are associated with a variety of disorders. Most of the disorders are part of metabolic syndromes e.g. impaired glucose tolerance, obesity, dyslipidemia and type 2 diabetes.
Studies have been undertaken on different ethnic groups showed positive association of a particular polymorphism of the adiponectin gene, SNP45 T > G with type 2 diabetes mellitus. Studies on Japanese by Nakatani et al. [4] and Hara et al. [8], on Europeans by Gable et al. [9], on Spanish population by González-Sánchez et al. [10], on Caucasian population by Menzaghi et al. [11], on the population of Uygurs of the Xinjiang region, China by Li et al. 2007 [12] support the association of SNP45 T > G with type 2 diabetes.
Impaired adiponectin multimerization due to mutation is usually the causative factor for the disease. Two mutants are incapable of forming the high molecular weight species while three are incapable of forming a stable trimer and show impaired secretion as well [13]. Thus the present study was aimed to find out the distribution of SNP45T > G of Adiponectin gene and its association with type 2 diabetes in selected population (patients attending OPD of Sri Ramachandra Hospital, Porur, Chennai).
Materials and Methods
This case control study was performed in SRMC Hospital in Chennai, Tamil Nadu, during the year 2007–2008. The study included 150 subjects in the age group of 40–70 years attending OPD of Sri Ramachandra Hospital, Porur Chennai. The sample size was selected following a similar study done on the population of Uygurs of the Xinjiang region, China by Li et al. [12]. The control group and the cases consist of 75 subjects each (age and sex matched). Although a post hoc calculation of the P value by Students’ t-test showed the power of this study to be <80%, but due to time and fund constraint, the sample size could not be increased. The project was approved by institutional ethical committee. The purpose of the study was explained in details to the participants and they had to sign a consent form. Controls were individuals with no clinically significant abnormal physical findings. They had normal clinically acceptable ECG, normal blood pressure and heart rate. BMI were in the range of 19–29 kg/m2. Moreover, all the control subjects had normal fasting (<110 mg/dl) and post prandial (<140 mg/dl) plasma glucose level. Existence of no other known illness was looked for in either group. All the subjects were able to communicate, competent and willing to give informed consent. The cases were clinically and biochemically confirmed as type 2 diabetes. Subjects having clinically significant neurological, cardiovascular, respiratory, endocrinal, gastrointestinal or other major systemic ailments, malignancy or undergoing major surgical procedures in past or suffering from any kind of acute illness were excluded from the study.
The data were collected through a standard questionnaire. All patients were interviewed regarding a full medical history that included age, sex, occupation, duration of diabetes, mode and duration of treatment, presence of any associated illness, surgical history, personal history of smoking/alcohol/drug abuse, dietary habit (vegetarian/nonvegetarian) and family history of diabetes.
About 5 ml of peripheral blood was collected from each subject in EDTA coated containers after receiving their consent. Samples were stored at −70°C till analysis.
DNA was isolated from the whole blood using standard salting out protocol. PCR analysis was done. Exon 2 of the adiponectin gene was amplified by PCR with specifically designed flanking primer sequences. Approximately 100 ng of genomic DNA was incubated in a total reaction volume of 20 μl, using 1.5 U Taq DNA polymerase (Bangalore Genei, India). The genotype frequency of SNP 45T > G was determined by using restriction enzyme Sma I (Bangalore Genei, India). The recognition site of the enzyme was CCC↓GGG. Concentration of the enzyme used in this study was (10 U/μl). Three kinds of genotypes: wild type TT, heterozygous type TG and homozygous variant type GG were studied by separating them on 2.5% agarose gel. The bands were visualized under UV transilluminator light. In each batch a few positive control samples for homozygous and heterozygous genotype were run to check repeatability. These positive controls were chosen from our previous study samples, and genotypes were confirmed by DNA sequencing analysis. Some samples were randomly genotyped by two different individuals to check sample replicability and found to be repeatable. In all about 20 per cent of study samples were analyzed in duplicate, with 100 per cent concordance in genotype calling. The protocols followed in the study were well standardized and performed with strict quality control measures.
Univariate and bivariate analysis were performed using SPSS 15. Pearson Chi-square test was performed to find the statistical significance between the genotypes.
Results
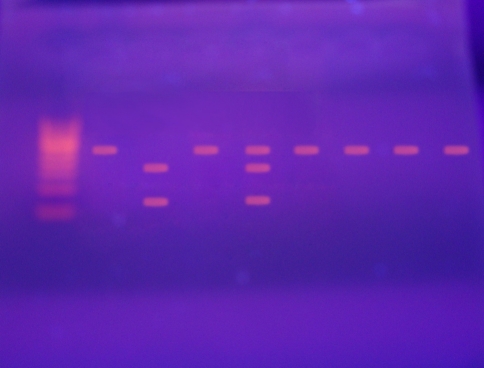
The purified PCR products underwent restriction digestion with Sma I. The following fragment sizing patterns were observed by agarose gel electrophoresis (Fig. 1).
Wild type 45TT: No cleavage of the whole 470 bp segment by Sma I.
Heterozygous variant type 45TG: Sma I cut at CCC↓GGG sequence to show three fragments in agarose gel electrophoresis (470 bp, 336 bp and 134 bp).
Homozygous variant type 45GG: Sma I cut at CCC↓GGG sequence to show two fragments in agarose gel electrophoresis (336 bp and 134 bp). This genotype was found only in one control subject.
Fig. 1.
Agarose gel electrophoresis showing the PCR–RFLP of adiponectin SNP45. Bands obtained Wild type TT (470 bp), heterozygous type TG (470 bp, 336 bp, 134 bp), homozygous mutant type GG (336 bp, 134 bp)
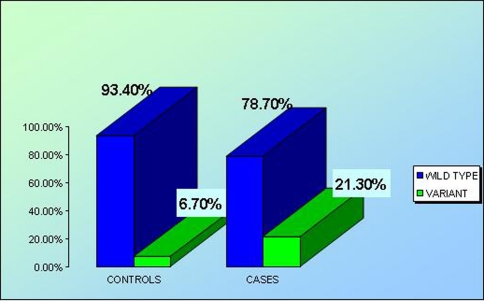
Total number and percentage of wild type and variants were calculated among cases and controls, tabulated in Table 1 and plotted in Fig. 2. The genotype frequencies of SNP 45 T > G of adiponectin gene of both case and control groups were calculated. Pearson Chi-square test was applied to determine the association between the variant form and occurrence of type 2 diabetes. The variant forms TG and GG are found to be significantly associated with type 2 diabetes mellitus (P < 0.01). Subjects carrying the variant types possess almost 4 times greater risk (Odd’s ratio = 3.80) of having type 2 diabetes than those carrying the wild type.
Table 1.
Distribution of polymorphism
| Polymorphism | Controls (N = 75) (%) | Cases (N = 75) (%) | P value | OR | 95% CI |
|---|---|---|---|---|---|
| Wild type (TT) | 70 (93.3) | 59 (78.7) | 0.010 | 3.797 | 1.312–10.983 |
| Heterozygote + variant (TG + GG)a | 5 (6.7) | 16 (21.3) |
aAt least any one should be present
Fig. 2.
Distribution of wild type and heterozygous + variant polymorphism among controls and cases (P value = 0.010, 95% CI = 1.312–10.983, ODD’S ratio = 3.797)
According to Table 2, the distribution of adiponectin genotypes and allele frequencies shows that cases were in HW whereas the control samples did not follow Hardy–Weinberg. The set of case and control under the present study was listed according to demographic variables in Table 3. Among the controls, 42.7% was male and 57.3% was female. Among the cases, 40% was male and 60% was female. 34.7% of controls were aged >50 years and 65.3% were aged <50 years. In cases, these numbers are 66.7 and 33.3%, respectively. In controls, 6% have the family history of type 2 diabetes, while in cases 56% have positive family history. 21.3% of controls were vegetarian and 78.7% are non vegetarian, while in cases 29.3% were vegetarian and 70.7% are non vegetarian. 12% of control population and 14.7% of cases had smoking habit. Alcohol intake was among 8% of controls and 10.7% of cases. 64% of controls and 58.7% of cases had BMI of >25.
Table 2.
Distribution of adiponectin genotypes and allele frequencies
| Sample | Genotypes | Allele Frequency | P value | |||
|---|---|---|---|---|---|---|
| TT | TG | GG | T | G | ||
| Control (N = 75) | ||||||
| Observed | 70 | 4 | 1 | 0.99 | 0.11 | <0.05 |
| Expected | 69.1 | 5.8 | 0.1 | |||
| Cases (N = 75) | ||||||
| Observed | 59 | 16 | 0 | 0.06 | 0.04 | NS |
| Expected | 59.9 | 14.3 | 0.9 | |||
Table 3.
Demographic variables for controls and cases
| Demographic variables | Controls (N = 75) (%) | P | Cases (N = 75) (%) | P |
|---|---|---|---|---|
| Sex | ||||
| Male | 32 (42.7) | >0.05 | 30 (40) | >0.05 |
| Female | 43 (57.3) | 45 (60) | ||
| Age | ||||
| Mean Age = 47.6 years | ||||
| >50 years | 26 (34.7) | >0.05 | ||
| <50 years | 49 (65.3) | |||
| Mean Age = 54.6 years | ||||
| >50 years | 50 (66.7) | >0.05 | ||
| <50 years | 25 (33.3) | |||
| Family history of Type 2 Diabetes | ||||
| Yes | 6 (8) | >0.05 | 42 (56) | >0.05 |
| No | 69 (92) | 33 (44) | ||
| Diet | ||||
| Vegetarian | 16 (21.3) | >0.05 | 22 (29.3) | >0.05 |
| Nonvegetarian | 59 (78.7) | 53 (70.7) | ||
| Smoking | ||||
| Yes | 9 (12) | >0.05 | 11 (14.7) | >0.05 |
| No | 66 (88) | 64 (85.3) | ||
| Alcohol | ||||
| Yes | 6 (8) | >0.05 | 8 (10.7) | >0.05 |
| No | 69 (92) | 67 (89.3) | ||
| BMI | ||||
| >25 | 48 (64) | >0.05 | 44 (58.7) | >0.05 |
| <25 | 27 (36) | 31 (41.3) | ||
The distribution of polymorphism in controls and cases according to demographic variables has been listed in Table 4. 6.3% males and 7% females in control group had the G allele (either TG or GG). In cases, 23.3% males and 20% females had the G allele. Among the controls, 3.85% people aged >50 years, 0% with positive family history, 11.1% smokers, 0% alcoholics and 10.5% people with BMI >25 had G alleles. Among cases, 23.3% male, 20% female, 18% people aged >50 years, 23.8% with positive family history, 27.3% smokers, 2.5% alcoholics and 18.2% people with BMI >25 had the G allele. However, no statistical significance is found among the polymorphism and any of the demographic variables.
Table 4.
Distribution of polymorphism according to demographical variables
| Polymorphism | Controls (N = 75) (%) | Cases (N = 75) (%) | P value | ||
|---|---|---|---|---|---|
| Male | Female | Male | Female | ||
| Wild type Heterozygote +Variant |
30 (93.8) | 40 (93) | 23 (76.7) | 36 (80) | >0.05 |
| 2 (6.3) | 3 (7) | 7 (23.3) | 9 (20) | ||
| Age < 50 years | Age > 50 years | Age < 50 years | Age > 50 years | ||
|---|---|---|---|---|---|
| Wild type Heterozygote +Variant |
45 (91.8) | 25 (96.15) | 18 (72) | 41 (82) | >0.05 |
| 4 (8.2) | 1 (3.85) | 7 (28) | 9 (18) |
| Family history | Family history | Family history | Family history | ||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| Wild type Heterozygote +Variant |
6 (100) | 64 (92.8) | 32 (76.2) | 27 (81.8) | >0.05 |
| 0 (0) | 5 (7.2) | 10 (23.8) | 6 (18.2) |
| Smoking + ve | Smoking − ve | Smoking + ve | Smoking − ve | ||
|---|---|---|---|---|---|
| Wild type Heterozygote +Variant |
8 (88.9) | 62 (93) | 8 (72.7) | 51 (79.7) | >0.05 |
| 1 (11.1) | 4 (7) | 3 (27.3) | 13 (20.3) |
| Alcohol + ve | Alcohol − ve | Alcohol + ve | Alcohol − ve | ||
|---|---|---|---|---|---|
| Wild type Heterozygote +Variant |
6 (100) | 64 (92.8) | 7 (87.5) | 52 (77.6) | >0.05 |
| 0 (0) | 5 (7.2) | 1 (2.5) | 15 (22.3) |
| BMI > 25 | BMI < 25 | BMI > 25 | BMI < 25 | ||
|---|---|---|---|---|---|
| Wild type Heterozygote + variant |
44 (89.5) | 26 (96.3) | 36 (81.8) | 23 (74.2) | >0.05 |
| 4 (10.5) | 1 (3.7) | 8 (18.2) | 8 (25.8) |
Discussion:
This result corroborates with findings of the studies on Japanese population by Nakatani et al. and Hara et al. [4, 8], on European population by Gable et al. [9], on Spanish population by González-Sánchez et al. [10], on Caucasian population by Menzaghi et al. [11], in the DESIR study group by Fumeron et al. [2], on the population of Uygurs of the Xinjiang region, China by Li et al. [12] and in the STOP-NIDDM trial by Zacharova et al. [14] .
In the STOP-NIDDM trial by Zacharova et al. [14], the G-allele of SNP 45 was associated with a 1.8-fold risk for type 2 diabetes. The present study correlates with the result of the STOP-NIDDM trial. The haplotypes containing G allele, i.e. TG and GG is significantly associated with a 3.797 fold increase for type 2 diabetes, if any one of them is present. Association of TG haplotype with type 2 diabetes correlates with report of Menzaghi et al. [11] based on the study on Caucasian subjects.
Nakatani et al. [4] in 2005 showed association of two single-nucleotide polymorphisms (SNP45T > G, SNP276G > T) with type 2 diabetes in the Japanese population. In their study, SNP45 was associated with insulin sensitivity (determined by HOMA-IR, P = 0.046) and obesity (body mass index; BMI, P = 0.043). SNP276 showed a stronger association with HOMA-IR (P = 0.018) and BMI (P = 0.017) in that study. Carriers with SNP45G-SNP276G haplotype had higher BMI (P = 0.034) and carriers with SNP45T-SNP276T haplotype had lower BMI (P = 0.005) and HOMA-IR (P = 0.037).
Hara et al. [8] in 2002 presented evidence of an association between frequent single nucleotide polymorphisms at positions 45 and 276 in the adiponectin gene and type 2 diabetes (P = 0.003 and P = 0.002, respectively). Subjects with the GG genotype at position 45 or the GG genotype at position 276 had a significantly increased risk of type 2 diabetes (odds ratio 1.70 [1.09–2.65] and 2.16 [1.22–3.95], respectively) compared with those having the TT genotype at positions 45 and 276, respectively. Here only GG haplotype is associated with insulin resistance. Neither homozygous nor heterozygous carriers of TG haplotype had an association with insulin sensitivity in the Japanese population.
Gable et al. [9] determined the impact of identified adiponectin gene (ADIPOQ) variants (−11391G > A, −1377C > G [promoter] and 45T > G [exon 2] and 276G > T [intron 2]) on the prospective risk of coronary artery disease and type 2 diabetes in healthy men. Only the +45T > G variant (3.80 [1.76–8.24]) was found to be associated with type 2 diabetes mellitus.
Result of the present study directly contradicts the study on Korean population as reported by Lee et al. [15]. They genotyped 427 non-diabetic controls and 493 type 2 diabetic patients for SNPs 45T > G and 276G > T of adiponectin gene and also measured plasma adiponectin concentrations in Koreans. No statistically significant differences were found in allele frequencies of SNPs 45 and 276 comparing control with type 2 diabetic subjects (T frequency 68.3 vs. 71.6%, P = 0.13 for SNP45, G frequency 72.2 vs. 68.9%, P = 0.12 for SNP276). They concluded that 45T > G and 276G > T of the adiponectin gene may not be important determinant of type 2 diabetes or insulin resistance in Korean subjects.
In addition, Vozarova de Courten et al. [16] screened the promoter, exons, and exon–intron boundaries of the adiponectin gene ACDC to identify allelic variants and to determine the role of genetic variation in ACDC in susceptibility to obesity and type 2 diabetes in Pima Indians. They identified 17 informative polymorphisms that comprised four common (minor allele frequency > 15%) linkage disequilibrium clusters consisting of 1–4 variants each. They genotyped one representative polymorphism from each cluster in 1,338 individuals and assessed genotypic association with type 2 diabetes, BMI, serum lipid levels, serum adiponectin levels, and measures of insulin sensitivity and secretion. None of the ACDC variants were associated with type 2 diabetes, BMI, or measures of insulin sensitivity or secretion. One variant, SNP-12823, was associated with serum adiponectin levels (P = 0.002), but this association explained only 2% of the variance of serum adiponectin levels. They concluded that that these common ACDC polymorphisms do not play a major role in susceptibility to obesity or type 2 diabetes in this population.
Study on Italians by Nannipieri et al. [17], French population based study by Vasseur F et al. [18] and Gu et al. [19] study report on Swedish subjects also showed no association among SNP45 T > G of adiponectin gene and insulin resistance. This may be due to variation in the ethnic group.
In the light of above data, adiponectin SNP45 T > G polymorphism may be closely correlated with the prevalence of type 2 diabetes mellitus. SNP45 T > G could trigger insulin resistance en route to the development of type 2 diabetes mellitus possibly through changes in mRNA stability, levels of adiponectin and eventually reduced plasma adiponectin concentrations. Thus we conclude that the presence of SNP45 T > G could be one of risk factors for developing type 2 diabetes mellitus.
The result of this study corroborates with the findings of several other studies performed on different ethnic groups. However, it contradicts some other studies. Therefore, the SNP45 T > G of adiponectin gene may play an important role in the development of type 2 diabetes mellitus. Direct evidence to support SNP45-induced regulation of adiponectin expression is still lacking and warrants further investigation. Direct measurement of adiponectin expression in white adipose tissues obtained from individuals with the SNP45 genotypes and its association with plasma fasting glucose level, insulin level, homeostasis model assessment (HOMA) for insulin resistance can throw more light on the role of SNP45 in the occurrence of type 2 diabetes mellitus.
References
- 1.Mohan V, Sandeep S, Deepa R, Shah B, Varghese C. Epidemiology of type 2 diabetes: Indian scenario. Indian J Med Res. 2007;125:217–230. [PubMed] [Google Scholar]
- 2.Vaxillaire M, Veslot J, Dina C, et al. Impact of common Type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes. 2008;57:244–254. doi: 10.2337/db07-0615. [DOI] [PubMed] [Google Scholar]
- 3.Vasseur F, Leprêtre F, Lacquemant C, et al. The genetics of adiponectin. Curr Diabetes. 2003;3(2):151–158. doi: 10.1007/s11892-003-0039-4. [DOI] [PubMed] [Google Scholar]
- 4.Nakatani K, Noma K, Nishioka J, et al. Adiponectin gene variation associates with the increasing risk of Type 2 diabetes in non-diabetic Japanese subjects. Int J Mol Med. 2005;15:173–177. [PubMed] [Google Scholar]
- 5.Trujillo ME, Scherer PE. Adiponectin––journey from an adipocyte secretory protein to biomarker of the metabolic syndrome. J Intern Med. 2005;257(2):167–175. doi: 10.1111/j.1365-2796.2004.01426.x. [DOI] [PubMed] [Google Scholar]
- 6.Viengchareum S, Zennaro MC, Tallec LPL, et al. Brown adipocytes are novel site of expression, regulation of adiponectin, resistin. FEBS Lett. 2002;532:345–350. doi: 10.1016/S0014-5793(02)03697-9. [DOI] [PubMed] [Google Scholar]
- 7.Hotta K, Funahashi T, Bodkin NL, et al. Circulating concentrations of the adipocyte protein adiponectin are decreased in parallel with reduced insulin sensitivity during the progression to type 2 diabetes in rhesus monkeys. Diabetes. 2001;50:1126–1133. doi: 10.2337/diabetes.50.5.1126. [DOI] [PubMed] [Google Scholar]
- 8.Hara K, Boutin P, Mori Y, et al. Genetic variation in the gene encoding adiponectin is associated with an increased risk of Type 2 diabetes in the Japanese population. Diabetes. 2002;51:536–540. doi: 10.2337/diabetes.51.2.536. [DOI] [PubMed] [Google Scholar]
- 9.Gable DR, Matin J, Whittall R, et al. Common adiponectin gene variants show different effects on risk of cardiovascular disease and Type 2 diabetes in European subjects. Ann Hum Genet. 2007;71(4):453–466. doi: 10.1111/j.1469-1809.2006.00340.x. [DOI] [PubMed] [Google Scholar]
- 10.González-Sánchez JL, Zabena CA, Martínez-Larrad MT, et al. An SNP in the adiponectin gene is associated with decreased serum adiponectin levels and risk for impaired glucose tolerance. Obes Res. 2005;13:807–812. doi: 10.1038/oby.2005.91. [DOI] [PubMed] [Google Scholar]
- 11.Menzaghi C, Ercolino T, Paola R, et al. As other features of the insulin resistance syndrome. Diabetes. 2002;51:2306–2312. doi: 10.2337/diabetes.51.7.2306. [DOI] [PubMed] [Google Scholar]
- 12.Li LL, Kang XL, Ran XJ, et al. Associations between 45T/G polymorphism of the adiponectin gene and plasma adiponectin levels with type 2 diabetes. Clin Exp Pharmacol Physiol. 2007;34(12):1287–1290. doi: 10.1111/j.1440-1681.2007.04713.x. [DOI] [PubMed] [Google Scholar]
- 13.Lyon CJ, Law RE, Hsueh WA. Minireview: adiposity, inflammation, and atherogenesis. Endocrinology. 2003;144:2195–2200. doi: 10.1210/en.2003-0285. [DOI] [PubMed] [Google Scholar]
- 14.Zacharova J, Chiasson JL, Laakso M, The STOP-NIDDM Study Group The common polymorphisms (single nucleotide polymorphism SNP +45 and SNP +276) of the adiponectin gene predict the conversion from impaired glucose tolerance to Type 2 Diabetes. The STOP-NIDDM trial. Diabetes. 2005;54:893–899. doi: 10.2337/diabetes.54.3.893. [DOI] [PubMed] [Google Scholar]
- 15.Lee YY, Lee NS, Cho YM, et al. Genetic association study of adiponectin polymorphisms with risk of Type 2 diabetes mellitus in Korean population. Diabet Med. 2005;22(5):569–575. doi: 10.1111/j.1464-5491.2005.01460.x. [DOI] [PubMed] [Google Scholar]
- 16.Vozarova de Courten B, Hanson RL, et al. Common polymorphisms in the adiponectin gene ACDC are not associated with diabetes in Pima Indians. Diabetes. 2005;54:284–289. doi: 10.2337/diabetes.54.1.284. [DOI] [PubMed] [Google Scholar]
- 17.Nannipieri M, Posadas R, Bonotti A, et al. Polymorphism of the 3′-untranslated region of the leptin receptor gene, but not the adiponectin SNP45 polymorphism, predicts Type 2 diabetes. Diabetes Care. 2006;29:2509–2511. doi: 10.2337/dc06-0355. [DOI] [PubMed] [Google Scholar]
- 18.Vasseur F, Helbecque N, Dina C, et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11(21):2607–2614. doi: 10.1093/hmg/11.21.2607. [DOI] [PubMed] [Google Scholar]
- 19.Gu HF, Abulaiti A, Östenson CG, et al. Single nucleotide polymorphisms in the proximal promoter region of the adiponectin (APM1) gene are associated with type 2 diabetes in Swedish Caucasians. Diabetes. 2004;53(1):S31–S35. doi: 10.2337/diabetes.53.2007.S31. [DOI] [PubMed] [Google Scholar]