FIG. 2.
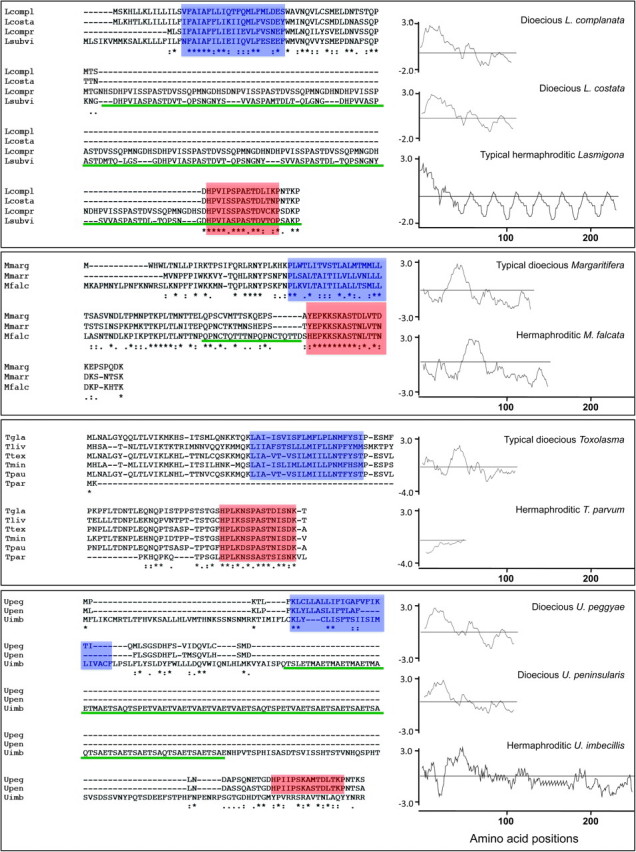
Primary and secondary structures of F-ORF and H-ORF amino acid sequences in dioecious and hermaphroditic freshwater mussel species, respectively. From above to below: Comparisons of dioecious Lasmigona spp. and hermaphroditic L. compressa and L. subviridis F- and H-ORFs. (Lcompl = L. complanata; Lcosta = L. costata; Lcompr = L. compressa; Lsubvi = L. subviridis). F-ORFs and H-ORF for Margaritifera margaritifera, M. marrianae and the hermaphroditic M. falcata. (Mmarg = M. margaritifera; Mmarr = M. marrianae; Mfalc = M. falcata). Comparisons of dioecious Toxolasma spp. and hermaphroditic T. parvum F-ORFs and H-ORF, respectively. (Tgla = T. glans; Tliv = T. lividus; Ttex = T. texasensis; Tmin = T. minor; Tpau = T. paulus; Tpar = T. parvum). F-ORFs and H-ORF for dioecious Utterbackia peggyae, U. peninsularis and hermaphroditic U. imbecillis. (Upeg = U. peggyae; Upen = U. peninsularis; Uimb = U. imbecillis). Alignment of amino acid sequences and secondary structures for the F- and H-ORFs are shown on the left. Color highlighting is as follows: blue box, the predicted transmembrane domain (TMH); reddish orange box, the relatively conserved, near-C-terminus, 15 amino acid motif; green, repetitive units. Hydropathy profiles are shown on the right. Numbers below profiles designate amino acid positions in each protein.
