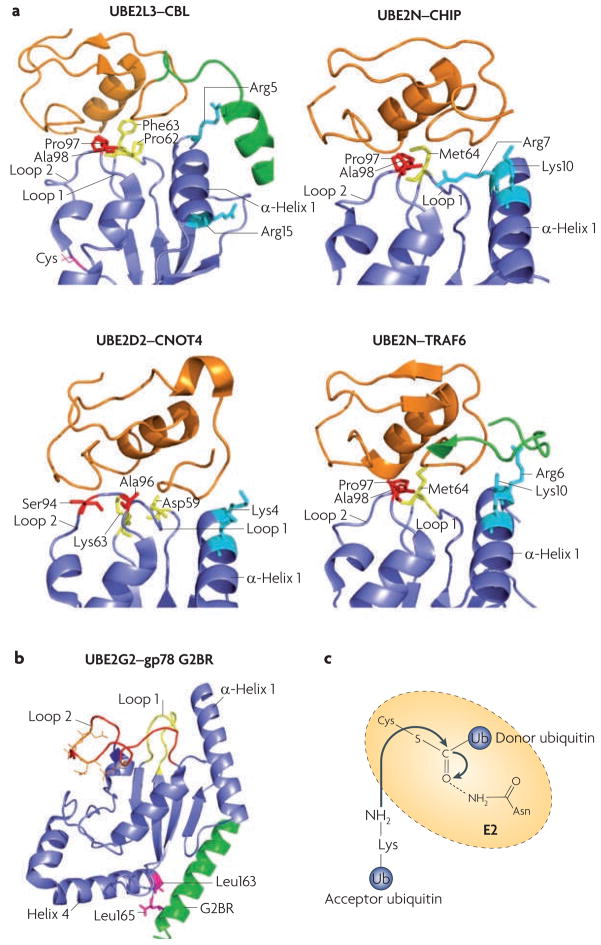
Figure 3. E2–E3 interaction specificity and the proposed mechanism of E2 catalysis.
a. Comparison of different ubiquitin-conjugating enzyme (E2)–ubiquitin ligase (E3) interactions: UBE2L3–casitas B-lineage lymphoma (CBL) (Protein Data Bank (PDB) code 1FBV44), UBE2N–carboxy terminus of HSC70 interacting protein (CHIP; also known as STUB1) (PDB code 2C2V46), UBE2D2–CNOT4 (CCR4–NOT transcription complex, subunit 4) (PDB code 1UR645) and UBE2N–TNF receptor associated factor 6 (TRAF6) (PDB code 3HCT47). All E2s use loop 1, loop 2 and α-helix 1 to interact with E3s. The side chains of the residues involved in E3 recognition are colour-coded (residues in loop 1 are yellow, in loop 2 are red and in α-helix 1 are cyan). Note that for CBL and TRAF6, E3 elements outside the RING domain (shown in green) participate in E2 binding. The CNOT4–UE2D2 structure is an NMR model, whereas the others were solved by crystallography. The RING finger (or the U-Box for CHIP) is in orange, and the cognate E2s are in blue. b. An example structure of a RING-independent E2–E3 interaction: that between UBE2G2 (blue) and the G2BR peptide from gp78 (also known as AMFR; green) (PBD code 3FSH)51. The E3 peptide interacts with two Leu residues of the E2, at a position remote from the predicted RING binding site (loop 1, loop 2 and α-helix 1). Acidic residues in the UBE2G2 acidic loop are in orange. c. A catalysis model for isopeptide bond formation during ubiquitin chain synthesis. The side chain of a conserved Asn residue in the E2 is proposed to interact with the active-site Cys (carrying the donor ubiquitin), which stabilizes the oxyanion transition state of the nucleophilic attack by the Lys residue of the acceptor ubiquitin.