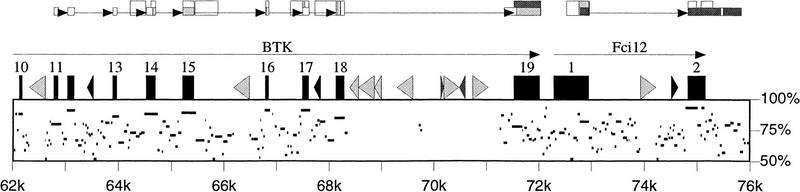
Figure 1.
Graphic representation of a genomic alignment together with EST matches computed by blEST. Human genes and interspersed repeats are drawn along the top of the box, with horizontal lines inside the box indicating the human positions and percent identity of gap-free portions of an alignment with the mouse genomic sequence. Above these are representations of the matches between the genomic sequences and the dbEST database (Boguski et al. 1993). The taller boxes show human ESTs matching the human sequence; and the shorter boxes show mouse ESTs matching the mouse sequence (with position on the human sequence deduced from the alignment). Shading of boxes indicates one match (white), two to three matches (light gray), or four to nine matches (dark gray). Thus, there are at least four mouse ESTs extending beyond the annotated end of the Fci12 gene, suggesting a longer 3′ UTR in the mouse. Arrows connecting EST boxes indicate introns identified by blEST using the sim4 strategy.