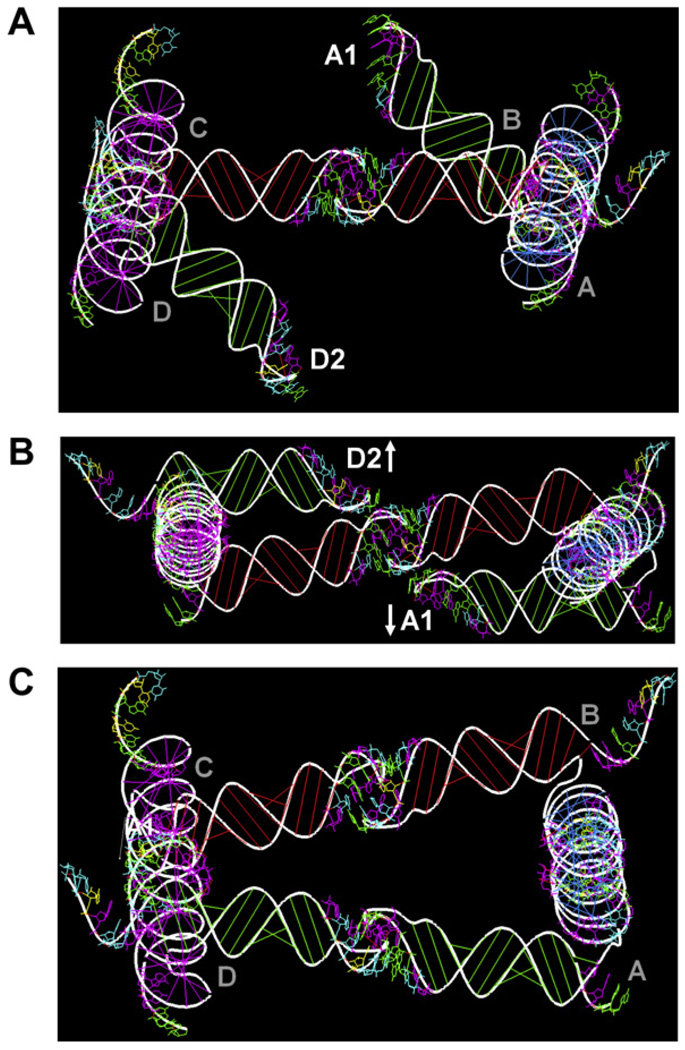
Fig. 3.
Modeling the LT17 type V tectosquare with RNA2D3D. The tectosquare is built out of four L-shaped monomers with the shaped H-loops and PDB-based RA motifs (1JJ2-50S ribosomal unit of Haloarcula marismortui). Full structure connectivity is specified in a topology script file. (A) The initial model does not close the tectosquare. (B) Adding 1 base pair to every 5′ ideal A-form helix of every L-shaped monomer induces effective rotation of every tectosquare side and causes the A1 and D2 H-loops to move past each other. (C) Co-axial rotation of each side (via the 5′ arm) by +22° brings the A1 and D2 H-loops into coaxial orientation and closes the full tectosquare structure. One L-shaped monomer modified in this way can then be used as a reference structure in search for the equivalent dynamic states of an unmodified monomer subjected to molecular dynamics simulation.