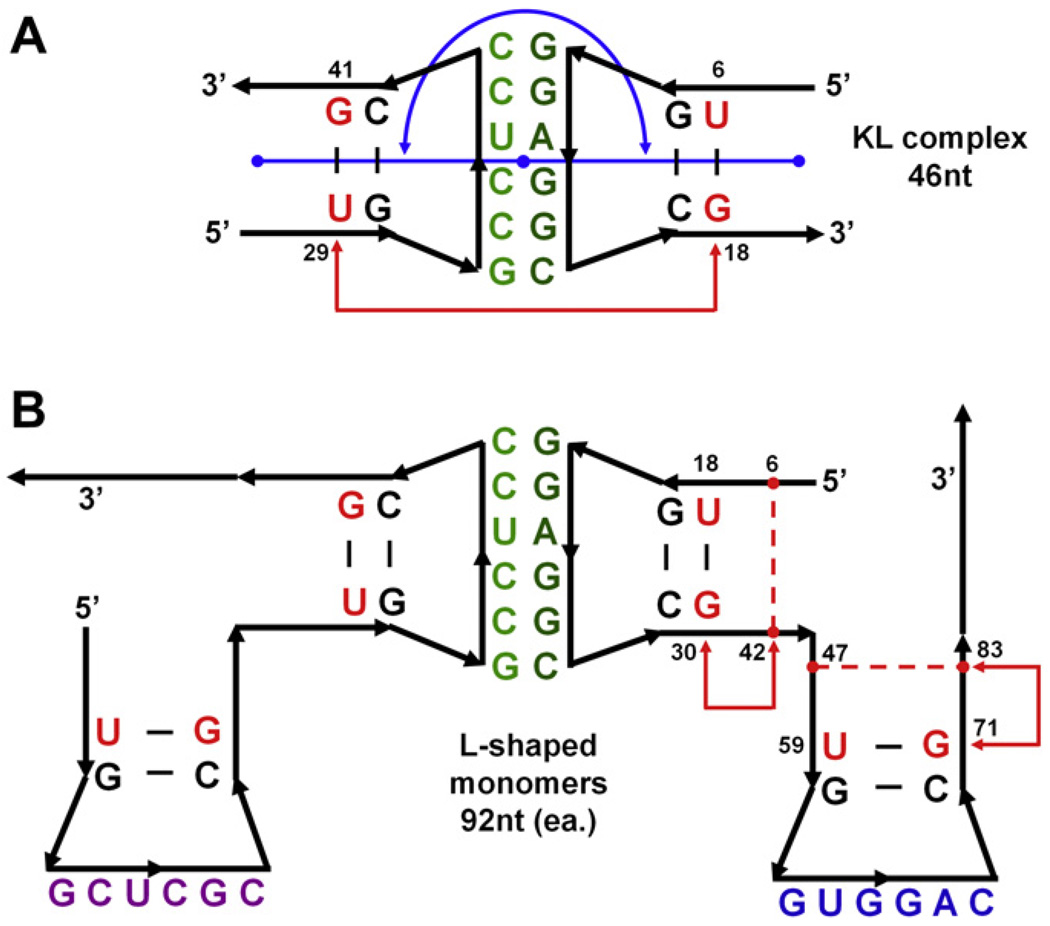
Fig. 4.
2D cartoon representations of tectosquare building blocks subjected to MD simulations. (A) Kissing loop complex, 46nt in two 23nt chains, mutated to match the A1D2 tectosquare interactions. Nucleotide labels indicate helix bases shared between the monomers (B) and kissing loop complexes. Bases highlighted in red indicate the nucleotides used as measuring points for the KL torsion angle (backbone P atoms of nucleotides 6, 18, 28 and 41 – refer to the text). The blue line and arc indicate the KL bending angle measured between the center of mass of the three base pairs at the end of each helix, with the mid-point at the center of mass of the six kissing loop base pairs, all represented with blue dots. (B) Two L-shaped monomers, each 92nt long (one was subjected to MD simulation). Nucleotides highlighted in red correspond to the red-labeled nucleotides in the KL complex (A) and were used as the L-KL interface points in combining the dynamic states of KLs and L-shaped monomers in searches for full structure closure. All the labeled nucleotides (red and black) within the L-shaped monomers and the KLs were used in chain-fitting of these blocks in PyMOL scripts, as described in the text. Red dashed lines and arrows within the two helical arms of the right monomer indicate positions used for the measurements of the helical torsion angles.