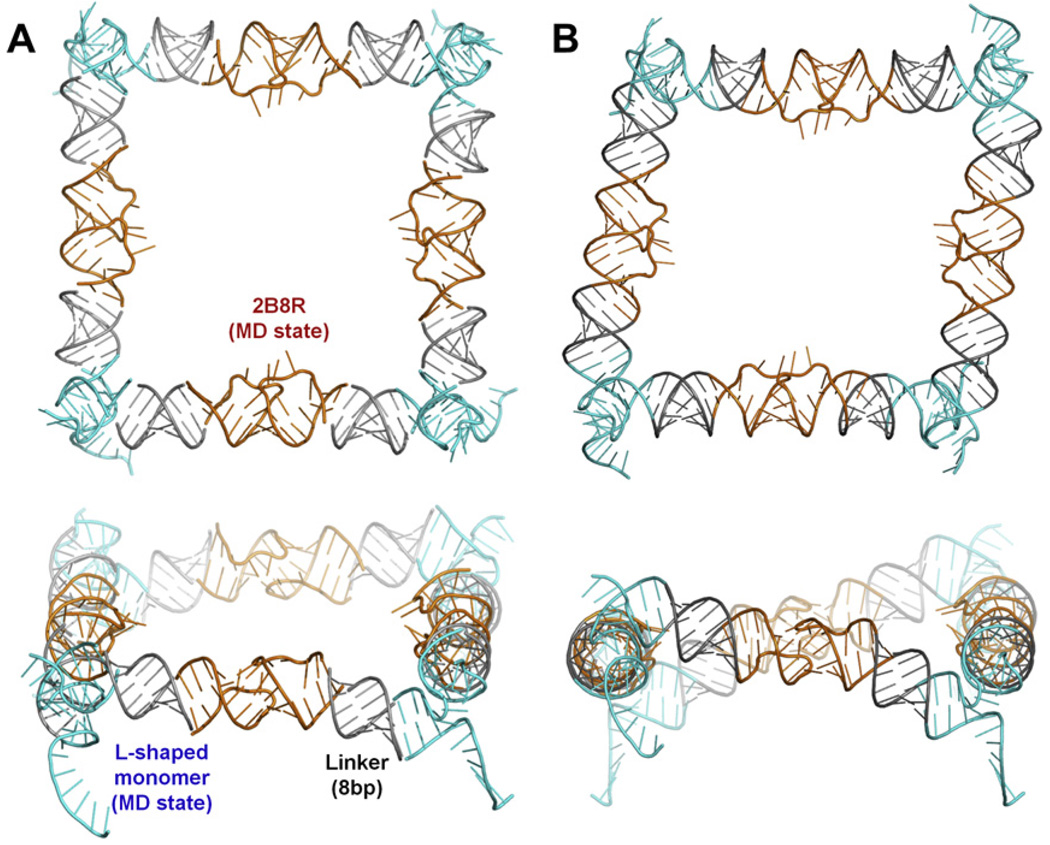
Fig. 9.
A closed tectosquare found in an automated NanoTiler search for KLs and L-shaped monomers linked with idealized helices to form the full structure closure. (A) The HIV-1 wild type kissing loop structure 2B8R (orange) was selected in the first round of searches where four copies of the 2B8R KLs were fit to four copies of idealized L-shaped monomers (created by RNA2D3D). The best scoring 2B8R MD state was then used in a search of the best tectosqure closing states in the L-shaped monomer MD trajectory. The monomers (cyan) were truncated to 4 base pairs in each arm and connected to the KLs via 8 bp long idealized helix linkers (gray). MD frame from the 7.6 ns point in the trajectory was selected (cyan). (B) The structure of the same combinations of the tectosquare building blocks as in A, after distortion of the linker helices performed by the NanoTiler to achieve the best fit, followed by the full structure energy minimization in Amber.