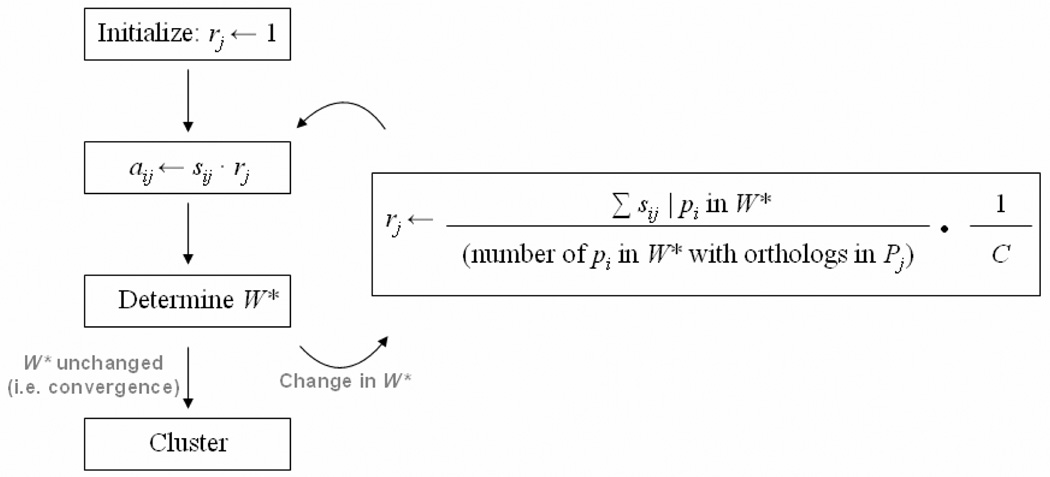
Figure 5.
A flow diagram of the APACE phylogenetic profile clustering method. The normalization factor, rj, is ultimately computed to be the multiplicand that equalizes the average similarity scores of every widely-conserved protein in P* to each reference proteome Pj and is initiated as 1. W* is the set of most conserved in P* and is by construction a strict subset of P*. The value of C is the arbitrary constant evolutionary distance every Pj is normalized to from P* through each rj.