Abstract
CD4+ T cells that selectively produce interleukin (IL)-17, are critical for host defense and autoimmunity1–4. Crucial for T helper17 (Th17) cells in vivo5,6, IL-23 has been thought to be incapable of driving initial differentiation. Rather, IL-6 and transforming growth factor (TGF)-β1 have been argued to be the factors responsible for initiating specification7–10. Herein, we show that Th17 differentiation can occur in the absence of TGF-β signaling. Neither IL-6 nor IL-23 alone efficiently generated Th17 cells; however, these cytokines in combination with IL-1β effectively induced IL-17 production in naïve precursors, independently of TGF-β. Epigenetic modification of the Il17a/Il17f and Rorc promoters proceeded without TGF-β1, allowing the generation of cells that co-expressed Rorγt and T-bet. T-bet+ Rorγt+ Th17 cells are generated in vivo during experimental allergic encephalomyelitis (EAE), and adoptively transferred Th17 cells generated with IL-23 without TGF-β1 were pathogenic in this disease model. These data suggest an alternative mode for Th17 differentiation. Consistent with genetic data linking IL23R with autoimmunity, our findings re-emphasize the importance of IL-23 and therefore have may have therapeutic implications.
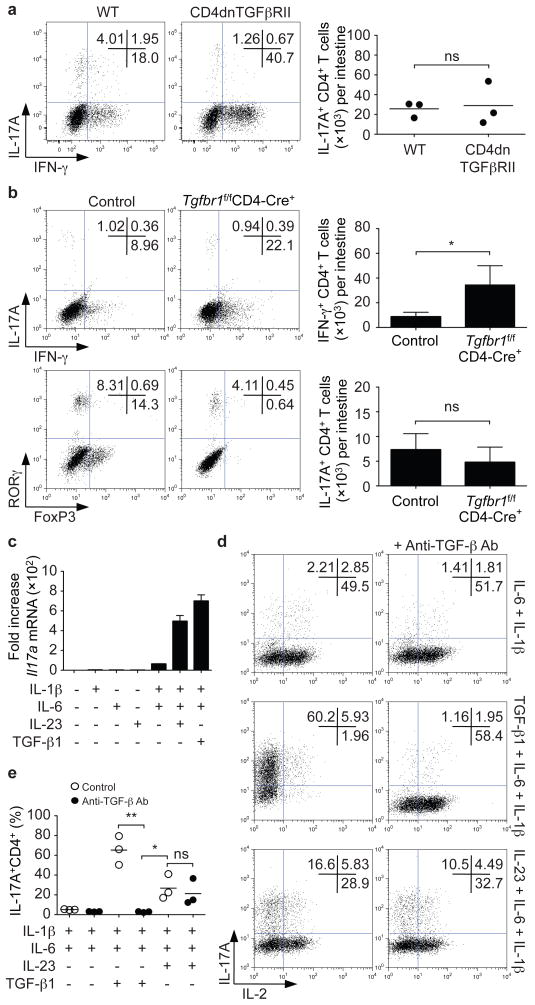
TGF-β1 is important for murine Th17 differentiation7,8 and accordingly, transgenic expression of a mutant TGF-β subunit receptor II (CD4dnTGFβRII) in T cells interferes with the generation of Th17 cells in the setting of EAE11. We revisited this issue by assessing whether Th17 cells were present in the intestinal lamina propria of these mice. As expected, increased proportions and absolute numbers of IFN-γ-producing CD4+ T cells were present (Supplementary Fig. 1a). Of note, IL-17-producing CD4+ cells were also present; the proportions were reduced, but there were no differences in the absolute numbers between wild type and CD4dnTGFβRII mice (Fig. 1a). We confirmed this finding using mice that lack TGF-β receptor subunit I in their T cells (Tgfbr1f/fCD4-Cre+ mice)12 (Fig. 1b). Rorγt-expressing CD4+ lamina propria T cells were also present in Tgfbr1f/fCD4-Cre+ mice, whereas Foxp3+CD4+ T cells were dramatically reduced (Fig. 1b). Collectively, these data argue that in vivo Th17 differentiation can occur in the absence of TGF-β signaling.
Figure 1. In vivo and in vitro differentiation of Th17 cells in the absence of TGF-β signaling.
a,b, Lamina propria cells were isolated from CD4dnTGFβRII and age-matched wild type (WT) mice (a) or Tgfbr1f/fCD4-Cre+ and Tgfbr1fl+CD4-Cre+ littermate controls (control) (b). Cells were stained for T cell markers and intracellular expression of IFN-γ, IL-17A, RORγt and FoxP3. Representative experiments are shown in left panels and pooled data are shown on the right (mean; error bars in b denote s.e.m., n=7). No significant differences in absolute numbers and proportions of IL-17A+CD4+ T cells were noted. *P<0.05. c–e, Naïve CD4+ T cells were isolated by cell sorting and activated in serum-free media with plate-bound anti-CD3/anti-CD28 for 4 days together with the indicated cytokines. Il17a mRNA expression was assessed by quantitative RT-PCR (c). IL-17A and IL-2 protein expression were analyzed by intracellular staining. Neutralizing anti-TGF-β antibodies prevented IL-6/IL-1β and TGF-β-dependent differentiation of Th17 cells, but not IL-23 and IL-6/IL-1β induced differentiation. Representative intracellular staining is depicted in panel d and pooled data from four individual experiments with mean values are shown in panel e. *P<0.05, **P<0.01.
We next explored whether it was possible to differentiate naïve Th17 cells in vitro in the absence of TGF-β. Sorted naïve CD4+ T cells cultured in serum-free medium with no exogenous cytokines failed to produce Il17a mRNA (Fig. 1c). The combination of IL-6, IL-1β and TGF-β1 efficiently induced Il17a mRNA, whereas IL-6 in the absence of TGF-β1 or IL-1β failed to do so. IL-23 alone failed to induce Il17a mRNA, but the combination of IL-6, IL-1β and IL-23 – without TGF-β – induced Il17a transcription (Fig. 1c) and protein expression (Fig. 1d and Supplementary Fig. 1b).
To exclude the possibility that endogenously produced TGF-β might contribute to Th17 differentiation, we cultured cells in the presence of anti-TGF-β antibodies. These antibodies blocked Th17 differentiation induced by TGF-β1/IL-6/IL-1β and enhanced IL-2-production but had no significant effect on Th17 differentiation induced by IL-23/IL-6/IL-1β (Fig. 1d, e). Similar results were also obtained in the absence of IL-1β and in medium supplemented with serum (Supplementary Fig. 1c, d). Anti-TGF-β antibodies blocked Foxp3-expression and Th17 differentiation over a wide concentration of exogenous TGF-β1, but had no effect on IL-23-dependent Th17 differentiation (Supplementary Fig. 2a). TGF-β-independent Th17 differentiation by IL-23/IL-6/IL-1β was also confirmed using a TGF-β receptor serine kinase inhibitor (TGFβRi) (Supplementary Fig. 2b), and T cells from CD4dnTGFβRII and Tgfbr1f/fCD4-Cre+ mice (Supplementary Fig. 2c, d). IL-23/IL-6/IL-1β-induced expression of Il21 and Il22 were also unaffected by the TGFβRi (Supplementary Fig. 2e). In fact, Th17 cells generated with IL-23/IL-6/IL-1β showed enhanced IL-22 production compared to TGF-β/IL-6/IL-1β-induced Th17 cells (Supplementary Fig. 2f). IL-17F expression was similar under both conditions. Absence of TGFβR1 expression and disruption of TGF-β signaling in Tgfbr1f/fCD4-Cre+ T cells or in T cells treated with the TGFβRi were confirmed by the absence of SMAD2 phosphorylation and gene induction by TGF-β1 (Supplementary Figs 2 g, h and 3 a, b).
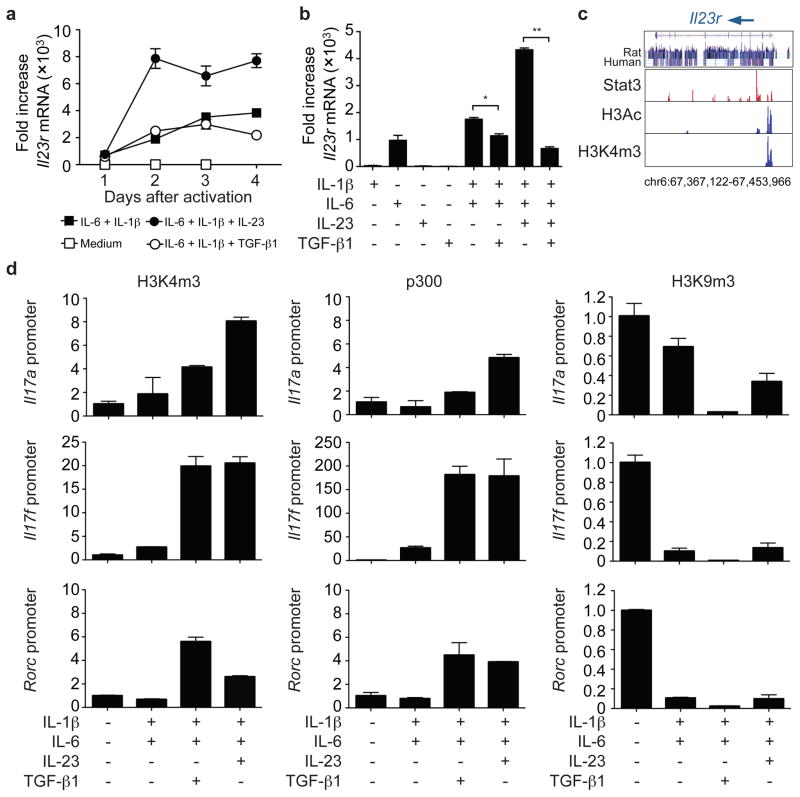
If our model is correct, an essential aspect of Th17 differentiation is the induction of IL-23R independently of TGF-β. We found that IL-6 and IL-1β induced Il23r mRNA expression in the absence of TGF-β. Addition of IL-23 significantly increased expression, whereas TGF-β1 inhibited expression (Fig. 2a, b, Supplementary Fig. 3c). Accordingly, the IL-23-mediated induction of the Th17 phenotype was altered by TGF-β1 (Supplementary Fig. 3d). The inhibitory effect of TGF-β signaling on Il23r expression was also evident using the TGFβRi and in cells with impaired TGF-β signaling (Supplementary Fig. 3e, f). Since IL-23 and IL-6 exert their effects through STAT3, we also documented that STAT3 binds the Il23r locus using chromatin immunoprecipitation with massive parallel sequencing (Fig. 2c).
Figure 2. IL-23 upregulates IL-23R and modifies the Il17 and Rorc loci in the absence of TGF-β.
a, b, Naïve CD4+ T cells were activated in serum-free media without cytokines, with individual cytokines or cytokine combinations as indicated. Il23r expression was analyzed by quantitative RT-PCR (mRNA levels ± s.e.m.) on days 1 to 4 after activation (a) or on day 4 only (b), *P<0.01, **P<0.001. c, IL-6 and IL-23 induce Stat3 binding to Il23r, histone 3 acetylation (H3Ac) and histone 3 lysine 4 trimethylation (H3K4m3) of the Il23r locus as determined by chromatin immunoprecipitation and massive parallel sequencing. d, Naïve CD4+ cells were activated as in (a). Fixed cells were immunoprecipitated with anti-H3K4m3, anti-p300 or anti-H3K9m3 antibodies. Eluted DNA was analyzed by quantitative PCR using primers spanning the promoter regions of Il17a, Il17f and Rorc.
We next determined if acquisition of permissive epigenetic modifications and extinction of repressive modifications of the Il17a/f and Rorc loci also occurred in the absence of TGF-β. In serum-free media, we found that in the presence of IL-6 and IL-1β, TGF-β and IL-23 were both effective in inducing histone 3 lysine 4 trimethylation (H3K4m3) of the Il17a and Il17f promoters (Fig. 2d). Recruitment of the transcriptional co-activator, p300, also occurred in response to both factors, whereas stimulation with IL-6 and IL-1β alone resulted in loss of the repressive mark, histone 3 lysine 9 trimethylation (H3K9m3). While TGF-β1 appeared to be a more efficient inducer of Rorc, p300 was recruited to this locus in response to IL-23 and Rorγt protein induction was detected in the absence of TGF-β1 (Fig. 2d and Supplementary Fig. 3g).
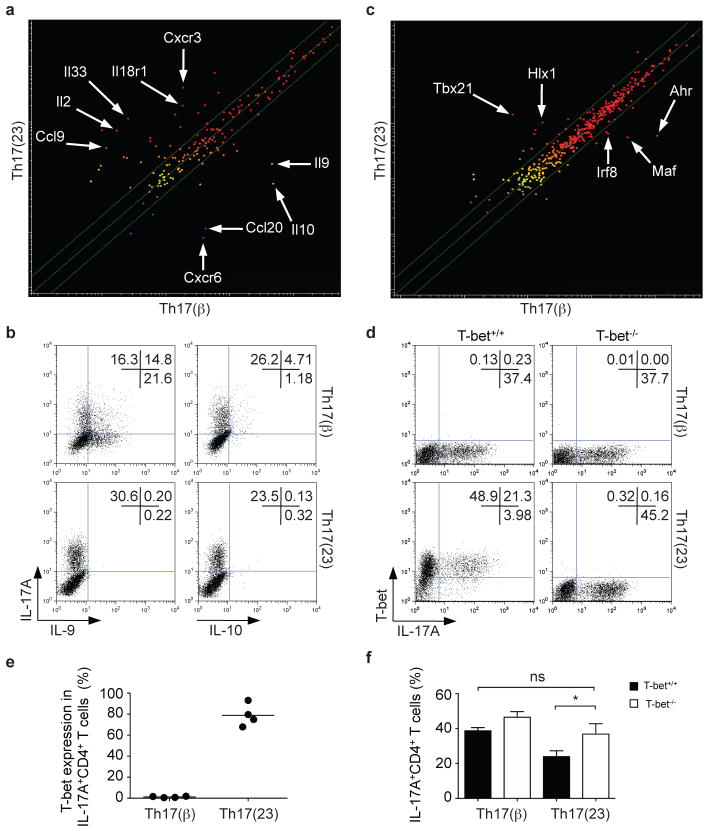
To compare IL-23/IL-6-induced Th17 [Th17(23)] cells with conventional Th17 cells [Th17(β)], we assessed global gene expression in both cells and found more than 2000 genes differentially expressed (Supplementary Fig. 4a, b). As shown in Fig. 3a, conventional Th17(β) cells expressed higher levels of Il9, Il10 and Ccl20, whereas Th17(23) cells expressed higher levels of Il2, Il33 and Il18r1. These differences were confirmed by quantitative PCR and measurement of protein expression (Fig. 3b, Supplementary Fig. 4 c–e). To assess whether IL-17-producing cells with a phenotype similar to those induced in vitro in the absence of TGF-β are generated in vivo, mice were immunized with ovalbumin and adjuvant. By day 7 the majority of IL-17-producing OT-II CD4+ T cells also expressed IL-18R1 and produced IL-26 (Supplementary Fig. 4f). Collectively, these data support the in vivo relevance of TGF-β-independent generation of Th17 cells.
Figure 3. IL-23-induced Th17 cells express T-bet but not IL-9 and IL-10.
a, Naïve CD4+ T cells were polyclonally stimulated in the presence of IL-6, IL-1β and either TGF-β (Th17(β) cells) or IL-23 (Th17(23) cells). Microarray analysis demonstrates the differential expression of genes encoding cytokines and receptors in the two subsets of Th17 cells. Mean values from two independent experiments are shown. b, Th17(β) and Th17(23) cells were polarized, expanded in IL-2, restimulated with anti-CD3/anti-CD28 antibodies (1 μg ml−1), cytokines, expanded and then analyzed for IL-17, IL-9 or IL-10 expression by intracellular staining. c, Transcription factor expression in Th17(β) and Th17(23) cells as assessed by microarray analysis. Mean values of two independent experiments are shown. d–f, Th17(23) but not Th17(β) express T-bet. Naïve CD4+ T cells were activated by IL-6, IL-1β with either IL-23 or TGF-β. IL-17 expression was not altered in T-bet−/− Th17(β) cells compared to T-bet+/+ Th17(β) cells. In contrast, loss of T-bet expression enhanced IL-17 production in Th17(23) cells. A representative experiment is depicted in d and pooled data are shown in e (n=4) and f (n=3, error bars are s.e.m., *P<0.05).
Conventional Th17(β) and IL-23-induced Th17(23) cells both expressed Rorc. As expected, Th17(β) cells expressed Maf and Ahr mRNA. In contrast, Th17(23) cells expressed Tbx21 and Hlx mRNA (Fig. 3c and Supplementary Fig. 5a, b) and T-bet protein (Figures 3d, e). In the absence of T-bet, the proportion of IL-17-producing cells induced by IL-23/IL-6/IL-1β was increased (Fig. 3f). Having demonstrated that Th17(23) cells retained T-bet expression, we wondered how this would influence cytokine production. As shown in Supplementary Fig. 6a, IL-23/IL-6/IL-1β induced slightly fewer IL-17-producing cells than the conventional pathway using TGF-β1. Expansion of cells in IL-2 and TGF-β1 resulted in reduction in IL-17 expression in both subsets. Th17 cells cultured in IL-12 acquired the ability to make IFN-γ13; however, this was significantly enhanced in Th17(23) cells. Previous studies have suggested that a major function of TGF-β is to downregulate T-bet and to prevent Th17 cells from acquiring the capacity to produce IFN-γ13,14. We therefore performed transcriptional profiling on conventional Th17 cells cultured with TGF-β1 and IL-6 in the presence and absence of the TGFβRi. In addition to contributing to the changes observed in cytokines and chemokines (Supplementary Fig. 5c), we found that blocking TGF-β1 resulted in the upregulation of Tbx21 and Gata3 mRNA (Supplementary Fig. 5a, d).
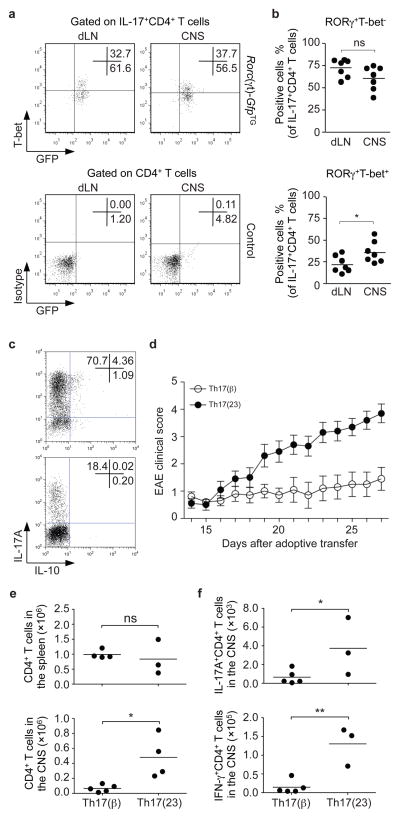
It is well established that IL-23 is critical for the development of EAE, but T-bet is also critical, a finding that does not fit well with our current understanding of Th17 cell development15. The present findings suggest that Th17 cells, which arise in the absence TGF-β, would express RORγt and T-bet. To test if such cells are seen in vivo, we immunized Rorc(γt)-GfpTG-reporter mice16 with myelin oligodendrocyte glycoprotein (MOG) 35–55 peptide. We found that approximately 25 to 60% of RORγt+ IL-17+ CD4+ cells in the CNS expressed T-bet (Fig. 4a, b). Furthermore, a significantly higher proportion of the RORγt, T-bet double positive Th17 cell population is found within the CNS compared with RORγt single positive Th17 cells (Fig. 4b).
Figure 4. RORγt+T-bet+ Th17 cells arise during CNS inflammation and T-bet-expressing, IL-23-induced Th17 cells are more pathogenic.
a, b, To induce CNS inflammation, we immunized Rorc(γt)-GfpTG or Rorc(γt)-GfpTG- control mice with MOG35–55 in CFA and CD4+ T cells isolated from the draining lymph nodes (dLN) or the CNS were analyzed by flow cytometry for IL-17, T-bet and GFP (RORγt) expression (upper panel). Lower panel shows isotype control staining for T-bet and fluorescence in transgene-negative littermates. A representative staining is depicted in a and pooled data of RORγt+ T-bet− and RORγt+ T-bet+ IL-17+CD4+ T cells are shown in b (n=7, *P<0.05). c, Naïve CD4+Vβ11+CD62L+CD44− were isolated by cell sorting from TCR(2D2) transgenic mice. The cells were activated with anti-CD3/anti-CD28, IL-6, IL-1β and anti-IFN-γ neutralizing antibodies with either TGF-β1 or IL-23 with anti-TGF-β neutralizing antibodies. The resultant cells were analyzed for IL-17 and IL-10 expression by intracellular staining and flow cytometry. d, Polarized cells (1×106) were adoptively transferred into Rag2−/− recipients and followed for signs of neurological disease. Data show mean ± s.e.m. of the EAE clinical score of 20 mice pooled from two independent experiments. e, f, CNS-infiltrating mononuclear cells were isolated and the total number of CD4+ T cells was determined in both groups (*P<0.01, e). The absolute numbers of CNS-infiltrating IL-17+ or IFN-γ+ CD4+ T cells of each group were assessed by intracellular cytokine staining (*P<0.05, **P<0.01, f).
To investigate the pathogenic potential of both subsets of Th17 cells, we isolated naïve T cells from TCR transgeneic (2D2) mice that recognize MOG35–55 peptide and differentiated them into Th17(23) cells in the presence of anti-TGF-β antibodies or alternatively into conventional Th17(β) cells resulting in higher percentage of IL-17-producing cells (Fig. 4c). After adoptive transfer, Th17(23) cells provoked significantly more severe disease than that induced by Th17(β) cells (Fig. 4d). This was associated with significantly greater total numbers of IL-17+ and IFN-γ T cells within the CNS (Fig. 4e, f) and elevated numbers of double-producer T cells (Supplementary Fig. 6c). In contrast, adoptive transfer of conventional Th17(β) cells were poorly pathogenic17 and preferentially trafficked to the spleen (Supplementary Fig. 6d).
Though initial reports suggested that IL-23 was a driver of IL-17 production2,18–20, the lack of IL-23R on naïve CD4+ T cells has been a factor in demoting this cytokine from an inducer of Th17 differentiation to a restricted role in Th17 expansion or preservation of pathogenicity6,21. In its place, TGF-β1 has been proposed to be the primary inductive factor that specifies Th17 differentiation in conjunction with IL-6. The current data indicate that TGF-β is not always necessary. IL-6 acting via STAT3 induces Il23r transcription, and IL-23 further enhances its own receptor expression. These cytokines in conjunction with IL-1β are sufficient to induce transcription and epigenetic modification of the Il17a/Il17f and Rorc loci, independently of TGF-β1. These data thus shed light on previous contradictory data pertaining to mouse versus human Th17 differentiation and indicate that there may be no species-specific difference22–24.
The present data do not dispute that idea that TGF-β1 and IL-6 induce Th17 differentiation (Supplementary Fig. 7). TGF-β is ubiquitous and undoubtedly is present at sites of inflammation. However, it is clear that TGF-β1 inhibits T-bet expression13,14. While this stabilizes the phenotype of selective IL-17 production, the Th17 cells generated in the presence of TGF-β1 and IL-6 are not pathogenic in the model tested and produce IL-9 and IL-1016,17,21,25,26. By contrast, Th17 cells derived in the presence of IL-23 appear to have greater pathogenic potential. They express T-bet and IL-18R1, which each are essential for the development of EAE15,17,27 and express CXCR3, which is important for trafficking of T cells to sites of inflammation28,29. In addition, TGF-β1 suppresses IL-23R expression and IL-23-mediated IL-22 production. The diversity of IL-17-producing cells helps to resolve previously perplexing data in EAE, namely the limited pathogenicity of Th17 cells generated with TGF-β and the importance of STAT3, IL-23R5,6,, T-bet15,17 and IL-18R127. The expression of characteristic “Th1” markers by Th17(23) cells underscores the complexity of Th cell lineage specification. Importantly, T-bet+Rorγt+ Th17 cells are present in lesional tissue in EAE, but also from patients with multiple sclerosis30. Thus, Th17 cells may represent heterogeneous populations comprising the offspring of Th17(β) or Th17(23) cells with distinct trafficking profiles and differing abilities to provoke autoimmune disease. These data are also of interest given the genetic link between IL23R polymorphisms and susceptibility to human autoimmune diseases. Future therapies for autoimmune disease should consider the phenotypic characters of pathogenic Th17 cells, generated in the absence of TGF-β, and their signaling pathways as possible targets.
METHODS SUMMARY
Mice
C57BL/6J, B6.Cg-Tg(Cd4-TGFBR2)16Flv/J (CD4dnTGFβRII), C57BL/6-Tg(Tcra2D2,Tcrb2D2)1Kuch/J (TCR(2D2)) and B6.129S6-Tbx21tm1Glm/J (T-bet−/−) mice were purchased from Jackson Laboratory (Bar Harbor, ME), B6.129S6-Rag2tm1Fwa (Rag2) mice from Taconic (Hudson, NY), Tgfbr1f/f mice were bred with CD4-Cre−transgenic mice and Tgfbr1f/+CD4-Cre+ littermates were used as controls. OT-II CD45.1+ mice, BAC-transgenic Rorc(γt)-GfpTG mice and Rorc(γt)-GfpTG− littermate controls were previously described6,16. All animal studies were performed according to the NIH guidelines for the use and care of live animals and were approved by the Institutional Animal Care and Use Committee of NIAMS or the IACUC of Schering-Plough Biopharma in accordance with guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care.
T cell isolation and differentiation
CD4+ T cells from spleens and lymph nodes of 6- to 8-week-old mice were purified by negative selection and magnetic separation (Miltenyi Biotec, Germany) followed by sorting of naive CD4+CD62L+CD44−CD25− population using FACSAria II (BD, NJ). Cells were activated by plate-bound anti-CD3/CD28 (both 10 μg ml−1; eBioscience, CA) in serum-free medium for 3–4 days either under neutral conditions or with IL-6 plus IL-1β(each 20 ng ml−1) and either human TGF-β1 (0.5 ng ml−1; Th17(β)) or IL-23 (50 ng ml−1; Th17(23)) (all from R&D Systems, MN). In all cell cultures anti-IFN-γ neutralizing antibodies (10 μg ml−1, BD Pharmingen) were added. IL-12 (PeproTech, NJ) was used at 20 ng ml−1, huIL-2 at 100 U ml−1. Procedures for antagonizing TGF-β-signaling, isolation of lamina propria lymphocytes and CNS-infiltrating lymphocytes are given in Methods.
Flow cytometry, real-time PCR, chromatin immunoprecipitation (ChIP), ChIP-seq, and microarray
Freshly isolated T cells or naïve T cells activated under the indicated conditions were analyzed. A detailed description of procedures and associated references are given in Methods.
Statistical analysis
For statistical analysis, all P values were calculated with Student’s t-test and P<0.05 was considered as significant.
Full Methods and associated references are available in the online version of the paper.
Supplementary Material
Acknowledgments
We thank Jim Simone, Jeff Lay (Flow Cytometry Section, NIAMS) and the NIAMS LACU staff for excellent technical support. This work has been supported by the Intramural Research Programs of NIAMS and NIAID.
Footnotes
Author Contributions
K.G. designed, performed, analyzed and interpreted all the experiments and wrote the manuscript. A.L., X.Y., M.J.G. and C.M.T. planned and performed experiments and helped to write the manuscript, L.W. and H.W.S. interpreted the microarray experiments and ChIP-seq data, H.L.R., W.T.W. and Y.K. performed and interpreted the ChIP-seq data, J.K., N.B., J.G. helped to analyze gut lymphocytes, T.D. and Q.C. helped to analyze CNS lymphocytes. G.E. provided the Rorc(γt)-GfpTG mice and made helpful suggestions, W.C. provided the Tgfbr1f/f mice, contributed to the experimental design and data interpretation, Y.B., E.S. and D.J.C. contributed to the experimental design, data interpretation and made helpful suggestions. J.O.S. contributed to the experimental design, analyzed and interpreted all acquired data and helped to write the manuscript.
The ChIP-seq and microarray data sets are deposited in Gene Expression Omnibus under accession numbers GSE23505 and GSEXXXXX.
References
- 1.Miossec P, Korn T, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med. 2009;361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 2.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 3.Stockinger B, Veldhoen M. Differentiation and function of Th17 T cells. Curr Opin Immunol. 2007;19:281–286. doi: 10.1016/j.coi.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 6.McGeachy MJ, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 9.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 10.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 11.Veldhoen M, Hocking RJ, Flavell RA, Stockinger B. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat Immunol. 2006;7:1151–1156. doi: 10.1038/ni1391. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, et al. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat Immunol. 2008;9:632–640. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- 13.Lee YK, et al. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das J, et al. Transforming growth factor beta is dispensable for the molecular orchestration of Th17 cell differentiation. J Exp Med. 2009;206:2407–2416. doi: 10.1084/jem.20082286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bettelli E, et al. Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis. J Exp Med. 2004;200:79–87. doi: 10.1084/jem.20031819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lochner M, et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgammat+ T cells. J Exp Med. 2008;205:1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, et al. T-bet is essential for encephalitogenicity of both Th1 and Th17 cells. J Exp Med. 2009;206:1549–1564. doi: 10.1084/jem.20082584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 19.Park H, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 21.McGeachy MJ, et al. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 22.Wilson NJ, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, Tato CM, Muul L, Laurence A, O’Shea JJ. Distinct regulation of interleukin-17 in human T helper lymphocytes. Arthritis Rheum. 2007;56:2936–2946. doi: 10.1002/art.22866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. 2007;8:942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 25.Elyaman W, et al. IL-9 induces differentiation of TH17 cells and enhances function of FoxP3+ natural regulatory T cells. Proc Natl Acad Sci U S A. 2009;106:12885–12890. doi: 10.1073/pnas.0812530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowak EC, et al. IL-9 as a mediator of Th17-driven inflammatory disease. J Exp Med. 2009;206:1653–1660. doi: 10.1084/jem.20090246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutcher I, Urich E, Wolter K, Prinz M, Becher B. Interleukin 18-independent engagement of interleukin 18 receptor-alpha is required for autoimmune inflammation. Nat Immunol. 2008;7:946–953. doi: 10.1038/ni1377. [DOI] [PubMed] [Google Scholar]
- 28.Lord GM, et al. T-bet is required for optimal proinflammatory CD4+ T-cell trafficking. Blood. 2005;106:3432–3439. doi: 10.1182/blood-2005-04-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch MA, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kebir H, et al. Preferential recruitment of interferon-gamma-expressing T(H)17 cells in multiple sclerosis. Ann Neurol. 2009;66:390–402. doi: 10.1002/ana.21748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.