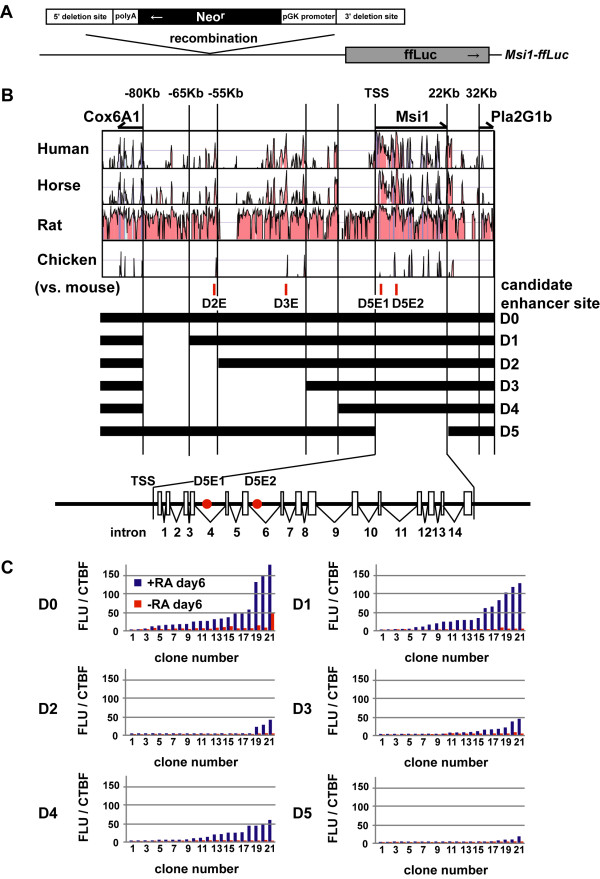
Figure 3.
Msi1 BAC deletion studies suggest that both the 55-65 kb 5' upstream region and the exon-intron coding region have Msi1 transcriptional activity. (A) Schematic representation of our deletion method. Targeted Msi1 enhancer/promoter regions were replaced with pgk pro.-Neor-bGH pA to select for successfully recombined clones. (B) The conserved genomic region between cox6a1 and pla2g1b in Msi1-ffLuc is shown. VISTA plots comparing the alignment of mouse versus human, horse, rat, and chicken. Full-length Msi1-ffLuc (D0) and deletion constructs (D1-D5) are also indicated. Blanks represent deleted regions replaced with pgk pro.-Neor-bGH pA. The positions of candidate enhancer sites used in later experiments are also shown (see details in Results). The exon-intron coding region of Msi1 gene is magnified. Red circles indicate candidate enhancer sites, white boxes indicate exons. (C) Gene expression in EBs derived from the deletion-reporter ES cell lines. All 21 cell lines were analysed for Luciferase expression in +RA neural induction cultures for 6 days (blue bars). Clones are presented in rank order for neural cultures. The red bars show the activity in the same clone under non-neural inducing conditions (-RA, not in rank order). Luciferase activity was normalized to the cell-viability fluorescence intensity using CellTiter-Blue (CTBF). The Msi1 expression in D0 and D1 was greatly increased under the neural-inducing conditions. Note that D5 showed decreased Msi1 expression; D2-D4, which included a 55-65-kb deletion, showed an even greater decrease.