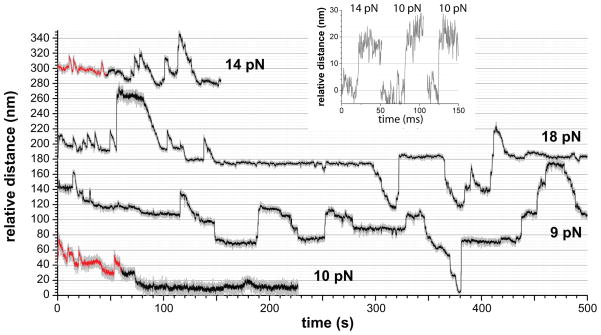
Figure 3. ClpX Unfolding and Translocation.
Experiments with ClpXSC alone showed some upward steps consistent with single-domain FLN unfolding and subsequent translocation (red regions). Highly cooperative unfolding transitions were observed (insert) in these regions. In the black regions, many upward steps were too large to represent single-domain unfolding, were frequently not followed by translocation, and probably correspond to load-induced backward slipping of substrate segments that had already been unfolded/translocated by ClpX. Following translocation through the ClpX pore, portions of the substrate may interact with the bead, refold, or be engaged by another ClpXSC enzyme, further complicating analysis. The combined forward and backward motions result in a slow rate of net tracking along the substrate (e.g., in both middle traces there is no net forward movement between 150 and 500 s). Mean velocities from Gaussian fits (± 95% confidence interval) of the distribution of translocation velocities for ClpXSC were 21.1 ± 2.9 aa s−1 at 4–8 pN and 18.3 ± 2.2 aa s−1 at 8–12 pN.