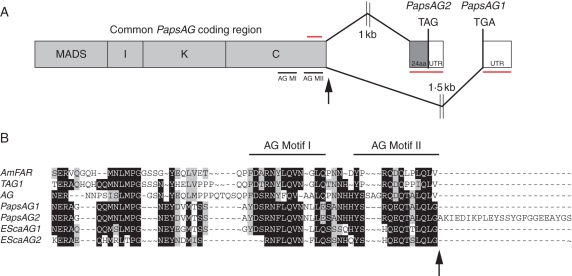
Fig. 1.
Gene structure of the Papaver somniferum AG gene. (A) Schematic showing how alternative splicing at the 3′ end of the coding sequence generates two transcripts, PapsAG-1 and PapsAG-2 (not drawn to scale). Regions underlined in red are gene-specific fragments used for gene-silencing with VIGS and for in situ mRNA hybridization probes. The region overlined with a red bar indicates the presence of a forward primer outside of the region used in vigs lines for expression analysis. Black arrows indicate the splicing donor site. AG MI, AG Motif I; AG MII, AG Motif II. (B) Alignment of the C-terminal regions of translated PapsAG-1 and PapsAG-2 sequences with AG orthologues from tomato (TAG; Q40168), Antirrhinum majus (AmFAR; CAB42988), Arabidopsis thaliana (AtAG; P17839) and Eschscholzia californica (EScaAG1 and EScaAG2; AAZ53205 and AAZ53206). PapsAG-2 has a 24-amino-acid extension. The conserved AG motifs are indicated.