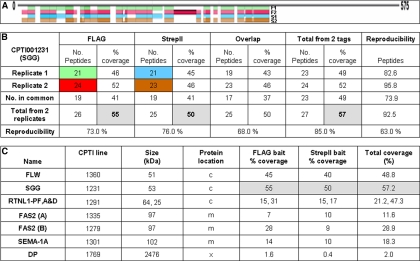
Fig. 3.
Identification, coverage and reproducibility of the bait protein in parallel purifications. A, Peptide coverage for FLAG and StrepII purified baits (SGG) from two replicates and (B) reproducibility of the Mascot processed MS identification data for SGG. Peptide reproducibility % calculated from (No. peptides in common/No. total from 2 reps) × 100. M = oxidation of Met residue. C, Summary of the combined replicate bait data for the six different proteins analyzed. More detailed analysis is in supplemental Table S2 online and supplemental Fig. S7 online. MS identified the presence of three different isoforms for Rtnl1: PA and PD (both 25 kDa) and PF (64 kDa) and the % sequence coverage are shown respectively. Two different protein trap lines were analyzed for Fas2 and the data listed separately as (A) and (B).