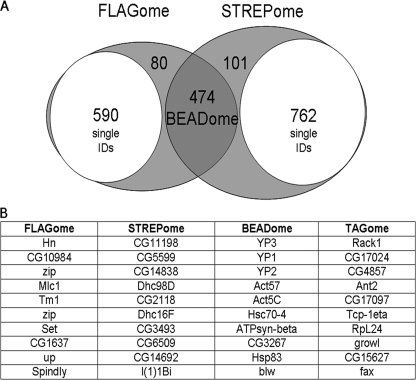
Fig. 4.
Analysis of negative control data. A, Multiple parallel affinity purifications of 25 nontagged lines (negative controls) and the use of ProteinCenter™ identified contaminants specific to FLAG and StrepII resins (FLAGome and STREPome respectively) and those common to both (BEADome). After identifying and removing the single protein hits (subsets) for each affinity method there is 72% overlap. B, The top ten hits for the FLAGome, STREPome and BEADome are listed in the table and the complete listings are presented in supplemental Tables S3A–S3C online. The top ten interacting proteins common to multiple baits are listed as the TRAPome and the complete list is presented in supplemental Table S5 online.