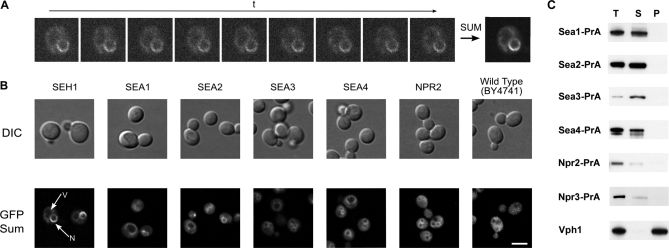
Fig. 5.
Proteins of the SEA complex are localized around vacuole membrane. Live florescence images of the SEA complex proteins genomically expressing GFP at their C terminus. A, Principle of the Sum Intensity Projection (SIP) algorithm, applied for localizing Seh1-GFP. Living cells were analyzed using a confocal-spinning disk microscope with low illumination power. Intensity values on a given pixel of the image are summed over all images in the time sequence to give the final image (right). B, Yeast cells were visualized by Nomarski optics (“DIC” row). GFP signals shown in the “GFP Sum” row were obtained by SIP or Maximum Intensity Projections of image sequences (duration or number of frames) taken with high exposure times (∼500 ms) to increase signal-to-noise ratio. Scale bar = 5 μm. C, Characterization of association of SEA complex proteins with enriched vacuole fraction. Total vacuole fractions (T) prepared from indicated PrA-tagged SEA complex proteins were treated with 0.1 m Na2CO3 prior to centrifugation at 100,000 × g. The resulting supernatant (S) and pellet (P) were analyzed by Western blotting with an IgG-HRP antibody. The distribution of vacuole integral membrane protein Vph1 was visualized with an anti-Vph1 antibody.